Gene Page: KPNA1
Summary ?
| GeneID | 3836 |
| Symbol | KPNA1 |
| Synonyms | IPOA5|NPI-1|RCH2|SRP1 |
| Description | karyopherin subunit alpha 1 |
| Reference | MIM:600686|HGNC:HGNC:6394|Ensembl:ENSG00000114030|HPRD:02819|Vega:OTTHUMG00000159487 |
| Gene type | protein-coding |
| Map location | 3q21 |
| Pascal p-value | 0.232 |
| Sherlock p-value | 0.095 |
| TADA p-value | 0.001 |
| Fetal beta | 0.922 |
| DMG | 1 (# studies) |
| eGene | Cerebellar Hemisphere Cerebellum |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| DNM:Girard_2011 | Whole Exome Sequencing analysis | The data set included 15 DNMs found from the exomes of 14 schizophrenia probands and their parents. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| KPNA1 | chr3 | 122146472 | C | A | NM_002264 NR_026698 | p.448E>* . | nonsense npcRNA | Schizophrenia | DNM:Girard_2011 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg00965985 | 3 | 122232605 | KPNA1 | 3.363E-4 | 0.387 | 0.041 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
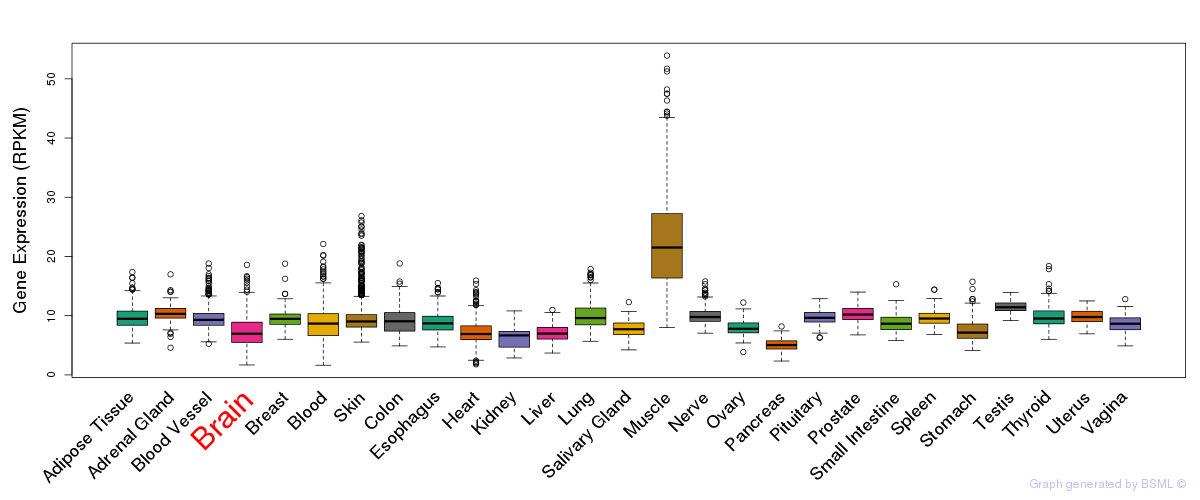
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CSE1L | CAS | CSE1 | MGC117283 | MGC130036 | MGC130037 | XPO2 | CSE1 chromosome segregation 1-like (yeast) | Reconstituted Complex | BioGRID | 10523667 |
| ELAVL1 | ELAV1 | HUR | Hua | MelG | ELAV (embryonic lethal, abnormal vision, Drosophila)-like 1 (Hu antigen R) | - | HPRD | 15342649 |
| HIST2H2BE | GL105 | H2B | H2B.1 | H2B/q | H2BFQ | MGC119802 | MGC119804 | MGC129733 | MGC129734 | histone cluster 2, H2be | - | HPRD | 11824786 |
| HIST3H3 | H3.4 | H3/g | H3FT | H3t | MGC126886 | MGC126888 | histone cluster 3, H3 | - | HPRD | 11824786 |
| ITK | EMT | LYK | MGC126257 | MGC126258 | PSCTK2 | IL2-inducible T-cell kinase | - | HPRD | 11581171 |
| KPNB1 | IMB1 | IPOB | Impnb | MGC2155 | MGC2156 | MGC2157 | NTF97 | karyopherin (importin) beta 1 | Reconstituted Complex | BioGRID | 9102465 |10523667 |
| LEF1 | DKFZp586H0919 | TCF1ALPHA | lymphoid enhancer-binding factor 1 | - | HPRD | 8631802 |
| LYAR | FLJ20425 | ZLYAR | Ly1 antibody reactive homolog (mouse) | Affinity Capture-MS | BioGRID | 17353931 |
| NUP54 | MGC13407 | nucleoporin 54kDa | Reconstituted Complex | BioGRID | 8707840 |
| NUP62 | DKFZp547L134 | FLJ20822 | FLJ43869 | IBSN | MGC841 | SNDI | p62 | nucleoporin 62kDa | Reconstituted Complex | BioGRID | 8707840 |
| PAX5 | BSAP | paired box 5 | - | HPRD | 10748034 |
| RAG1 | MGC43321 | RNF74 | recombination activating gene 1 | - | HPRD,BioGRID | 8052633 |
| RAG2 | RAG-2 | recombination activating gene 2 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 12861017 |
| RANBP2 | NUP358 | TRP1 | TRP2 | RAN binding protein 2 | Reconstituted Complex | BioGRID | 10473610 |
| RUNX1T1 | AML1T1 | CBFA2T1 | CDR | ETO | MGC2796 | MTG8 | MTG8b | ZMYND2 | runt-related transcription factor 1; translocated to, 1 (cyclin D-related) | - | HPRD,BioGRID | 10951564 |
| STAT1 | DKFZp686B04100 | ISGF-3 | STAT91 | signal transducer and activator of transcription 1, 91kDa | - | HPRD,BioGRID | 12048190 |
| UBR5 | DD5 | EDD | EDD1 | FLJ11310 | HYD | KIAA0896 | MGC57263 | ubiquitin protein ligase E3 component n-recognin 5 | - | HPRD,BioGRID | 12011095 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID PI3KCI AKT PATHWAY | 35 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | 66 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | 13 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERFERON SIGNALING | 159 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOSIS | 148 | 94 | All SZGR 2.0 genes in this pathway |
| REACTOME INFLUENZA LIFE CYCLE | 203 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV INFECTION | 207 | 122 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | 27 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME HOST INTERACTIONS OF HIV FACTORS | 132 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | 33 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOTIC EXECUTION PHASE | 54 | 37 | All SZGR 2.0 genes in this pathway |
| IGARASHI ATF4 TARGETS DN | 90 | 65 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN | 455 | 304 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 UP | 418 | 263 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA DN | 146 | 94 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HIF1A TARGETS UP | 67 | 40 | All SZGR 2.0 genes in this pathway |
| MAYBURD RESPONSE TO L663536 DN | 56 | 32 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| KONG E2F1 TARGETS | 10 | 6 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION DN | 517 | 309 | All SZGR 2.0 genes in this pathway |
| GUO HEX TARGETS DN | 65 | 36 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS LATE PROGENITOR | 544 | 307 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR DN | 354 | 216 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 UP | 344 | 215 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |