Gene Page: KRT15
Summary ?
| GeneID | 3866 |
| Symbol | KRT15 |
| Synonyms | CK15|K15|K1CO |
| Description | keratin 15 |
| Reference | MIM:148030|HGNC:HGNC:6421|HPRD:01009| |
| Gene type | protein-coding |
| Map location | 17q21.2 |
| Pascal p-value | 0.405 |
| TADA p-value | 0.011 |
| Fetal beta | -0.041 |
| eGene | Myers' cis & trans |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| KRT15 | chr17 | 39673416 | A | T | NM_002275 NM_002275 | . p.167I>N | splice missense | Schizophrenia | DNM:Fromer_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17799628 | chr9 | 106442995 | KRT15 | 3866 | 0.19 | trans |
Section II. Transcriptome annotation
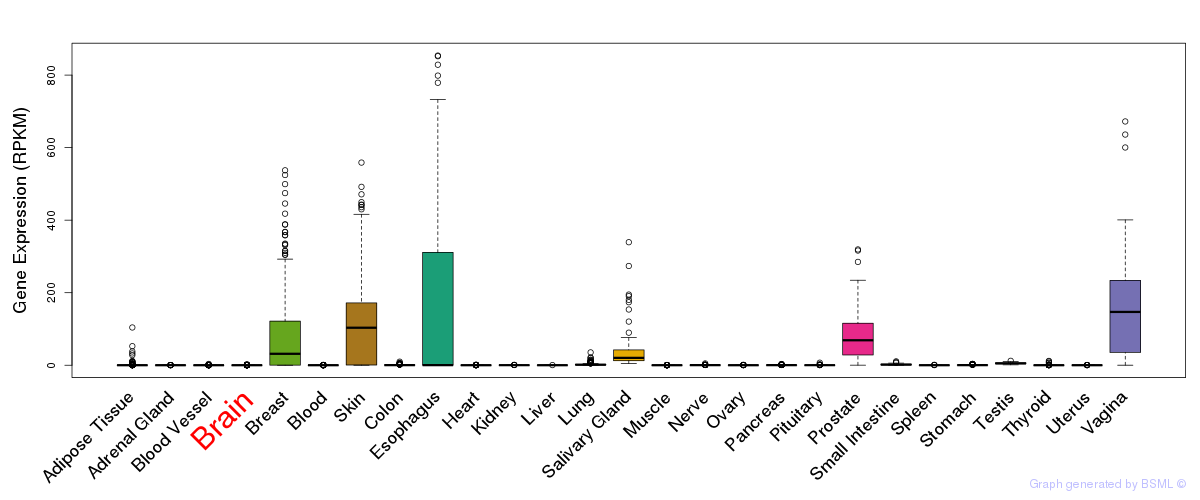
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABI2 | ABI-2 | ABI2B | AIP-1 | AblBP3 | SSH3BP2 | argBPIA | argBPIB | abl interactor 2 | Two-hybrid | BioGRID | 16189514 |
| ABI3 | NESH | SSH3BP3 | ABI family, member 3 | Two-hybrid | BioGRID | 16189514 |
| ABLIM1 | ABLIM | DKFZp781D0148 | FLJ14564 | KIAA0059 | LIMAB1 | LIMATIN | MGC1224 | actin binding LIM protein 1 | Two-hybrid | BioGRID | 16189514 |
| AMOTL2 | LCCP | angiomotin like 2 | Two-hybrid | BioGRID | 16189514 |
| ANKRD26L1 | MGC12538 | ankyrin repeat domain 26-like 1 | Two-hybrid | BioGRID | 16189514 |
| ARC | Arg3.1 | KIAA0278 | activity-regulated cytoskeleton-associated protein | Two-hybrid | BioGRID | 16189514 |
| C10orf10 | DEPP | FIG | chromosome 10 open reading frame 10 | Two-hybrid | BioGRID | 16189514 |
| C1orf216 | FLJ38984 | chromosome 1 open reading frame 216 | Two-hybrid | BioGRID | 16189514 |
| C1orf65 | FLJ35728 | RP11-76K24.1 | chromosome 1 open reading frame 65 | Two-hybrid | BioGRID | 16189514 |
| CALCOCO1 | Cocoa | KIAA1536 | PP13275 | calphoglin | calcium binding and coiled-coil domain 1 | Two-hybrid | BioGRID | 16189514 |
| CCDC101 | FLJ32446 | coiled-coil domain containing 101 | Two-hybrid | BioGRID | 16189514 |
| CCDC120 | JM11 | coiled-coil domain containing 120 | Two-hybrid | BioGRID | 16189514 |
| DGCR6L | FLJ10666 | DiGeorge syndrome critical region gene 6-like | Two-hybrid | BioGRID | 16189514 |
| EXOC8 | EXO84 | Exo84p | SEC84 | exocyst complex component 8 | Two-hybrid | BioGRID | 16189514 |
| FAM107A | DRR1 | FLJ30158 | FLJ45473 | TU3A | family with sequence similarity 107, member A | Two-hybrid | BioGRID | 16189514 |
| FAM164C | C14orf140 | FLJ23093 | family with sequence similarity 164, member C | Two-hybrid | BioGRID | 16189514 |
| HOXB6 | HOX2 | HOX2B | HU-2 | Hox-2.2 | homeobox B6 | Two-hybrid | BioGRID | 16189514 |
| KIAA0408 | FLJ43995 | RP3-403A15.2 | KIAA0408 | Two-hybrid | BioGRID | 16189514 |
| KIAA1217 | DKFZp761L0424 | MGC31990 | SKT | KIAA1217 | Two-hybrid | BioGRID | 16189514 |
| KIAA1267 | DKFZp686P06109 | DKFZp727C091 | MGC102843 | KIAA1267 | Two-hybrid | BioGRID | 16189514 |
| KRT18 | CYK18 | K18 | keratin 18 | Two-hybrid | BioGRID | 16189514 |
| KRT19 | CK19 | K19 | K1CS | MGC15366 | keratin 19 | Two-hybrid | BioGRID | 16189514 |
| KRT20 | CD20 | CK20 | K20 | KRT21 | MGC35423 | keratin 20 | Two-hybrid | BioGRID | 16189514 |
| KRT6A | CK6A | CK6C | CK6D | K6A | K6C | K6D | KRT6C | KRT6D | keratin 6A | Two-hybrid | BioGRID | 16189514 |
| KRT6B | CK6B | K6B | KRTL1 | PC2 | keratin 6B | Two-hybrid | BioGRID | 16189514 |
| KRT81 | HB1 | Hb-1 | KRTHB1 | MLN137 | ghHkb1 | hHAKB2-1 | keratin 81 | Two-hybrid | BioGRID | 16189514 |
| LDOC1 | BCUR1 | Mar7 | Mart7 | leucine zipper, down-regulated in cancer 1 | Two-hybrid | BioGRID | 16189514 |
| LNX1 | LNX | MPDZ | PDZRN2 | ligand of numb-protein X 1 | Two-hybrid | BioGRID | 16189514 |
| MAD2L1BP | CMT2 | KIAA0110 | MGC11282 | RP1-261G23.6 | MAD2L1 binding protein | Two-hybrid | BioGRID | 16189514 |
| MCM10 | CNA43 | DNA43 | MGC126776 | PRO2249 | minichromosome maintenance complex component 10 | Two-hybrid | BioGRID | 16189514 |
| NIF3L1 | ALS2CR1 | CALS-7 | MDS015 | NIF3 NGG1 interacting factor 3-like 1 (S. pombe) | Two-hybrid | BioGRID | 16189514 |
| NOC4L | MGC3162 | NOC4 | nucleolar complex associated 4 homolog (S. cerevisiae) | Two-hybrid | BioGRID | 16189514 |
| NUDT18 | FLJ22494 | nudix (nucleoside diphosphate linked moiety X)-type motif 18 | Two-hybrid | BioGRID | 16189514 |
| PCM1 | PTC4 | pericentriolar material 1 | Two-hybrid | BioGRID | 16189514 |
| PRPH | NEF4 | PRPH1 | peripherin | Two-hybrid | BioGRID | 16189514 |
| PSMA1 | HC2 | MGC14542 | MGC14575 | MGC14751 | MGC1667 | MGC21459 | MGC22853 | MGC23915 | NU | PROS30 | proteasome (prosome, macropain) subunit, alpha type, 1 | Two-hybrid | BioGRID | 16189514 |
| PSMC5 | S8 | SUG1 | TBP10 | TRIP1 | p45 | p45/SUG | proteasome (prosome, macropain) 26S subunit, ATPase, 5 | Two-hybrid | BioGRID | 16189514 |
| RCOR3 | FLJ10876 | FLJ16298 | RP11-318L16.1 | REST corepressor 3 | Affinity Capture-Western Two-hybrid | BioGRID | 16189514 |
| RHPN1 | KIAA1929 | ODF5 | RHOPHILIN | RHPN | rhophilin, Rho GTPase binding protein 1 | Two-hybrid | BioGRID | 16189514 |
| RIBC2 | C22orf11 | RIB43A domain with coiled-coils 2 | Two-hybrid | BioGRID | 16189514 |
| SMARCD1 | BAF60A | CRACD1 | Rsc6p | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 | Two-hybrid | BioGRID | 16189514 |
| TCHP | MGC10854 | trichoplein, keratin filament binding | Two-hybrid | BioGRID | 16189514 |
| TEKT4 | MGC27019 | tektin 4 | Two-hybrid | BioGRID | 16189514 |
| TSG101 | TSG10 | VPS23 | tumor susceptibility gene 101 | Two-hybrid | BioGRID | 16189514 |
| TUBGCP4 | 76P | FLJ14797 | GCP4 | tubulin, gamma complex associated protein 4 | Two-hybrid | BioGRID | 16189514 |
| TXNDC11 | EFP1 | thioredoxin domain containing 11 | Two-hybrid | BioGRID | 16189514 |
| USHBP1 | AIEBP | FLJ38709 | FLJ90681 | MCC2 | Usher syndrome 1C binding protein 1 | Two-hybrid | BioGRID | 16189514 |
| USP2 | UBP41 | USP9 | ubiquitin specific peptidase 2 | Affinity Capture-Western Two-hybrid | BioGRID | 16189514 |
| WAC | BM-016 | MGC10753 | PRO1741 | Wwp4 | bA48B24 | bA48B24.1 | WW domain containing adaptor with coiled-coil | Two-hybrid | BioGRID | 16189514 |
| ZNF638 | DKFZp686P1231 | MGC26130 | MGC90196 | NP220 | ZFML | Zfp638 | zinc finger protein 638 | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| LIU PROSTATE CANCER DN | 481 | 290 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST DUCTAL CARCINOMA VS DUCTAL NORMAL DN | 198 | 110 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST LOBULAR CARCINOMA VS DUCTAL NORMAL DN | 91 | 53 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST LOBULAR CARCINOMA VS LOBULAR NORMAL UP | 94 | 59 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN | 455 | 304 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER BASAL VS MESENCHYMAL UP | 121 | 45 | All SZGR 2.0 genes in this pathway |
| ROY WOUND BLOOD VESSEL DN | 22 | 14 | All SZGR 2.0 genes in this pathway |
| JAEGER METASTASIS DN | 258 | 141 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS DN | 232 | 139 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA UP | 171 | 112 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HIF1A TARGETS DN | 91 | 58 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HIF1A AND HIF2A TARGETS DN | 104 | 72 | All SZGR 2.0 genes in this pathway |
| LIU CDX2 TARGETS DN | 8 | 5 | All SZGR 2.0 genes in this pathway |
| COLDREN GEFITINIB RESISTANCE DN | 230 | 115 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP | 487 | 286 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION DN | 206 | 136 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN | 464 | 276 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 1 DN | 169 | 102 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 3 DN | 59 | 32 | All SZGR 2.0 genes in this pathway |
| YAO HOXA10 TARGETS VIA PROGESTERONE UP | 79 | 58 | All SZGR 2.0 genes in this pathway |
| LEI MYB TARGETS | 318 | 215 | All SZGR 2.0 genes in this pathway |
| HENDRICKS SMARCA4 TARGETS UP | 56 | 35 | All SZGR 2.0 genes in this pathway |
| CAMPS COLON CANCER COPY NUMBER UP | 92 | 45 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| HUPER BREAST BASAL VS LUMINAL DN | 59 | 44 | All SZGR 2.0 genes in this pathway |
| CROMER TUMORIGENESIS DN | 51 | 29 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS DN | 306 | 191 | All SZGR 2.0 genes in this pathway |
| YIH RESPONSE TO ARSENITE C5 | 10 | 7 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS NOT VIA AKT1 UP | 211 | 131 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS VIA AKT1 UP | 281 | 183 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL DN | 308 | 187 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |