Gene Page: MAPT
Summary ?
| GeneID | 4137 |
| Symbol | MAPT |
| Synonyms | DDPAC|FTDP-17|MAPTL|MSTD|MTBT1|MTBT2|PPND|PPP1R103|TAU |
| Description | microtubule associated protein tau |
| Reference | MIM:157140|HGNC:HGNC:6893|Ensembl:ENSG00000186868|HPRD:01142|Vega:OTTHUMG00000168833 |
| Gene type | protein-coding |
| Map location | 17q21.1 |
| Sherlock p-value | 0.004 |
| Fetal beta | -2.487 |
| eGene | Cerebellar Hemisphere Cerebellum Cortex Frontal Cortex BA9 Myers' cis & trans Meta |
| Support | STRUCTURAL PLASTICITY G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| CV:PheWAS | Phenome-wide association studies (PheWAS) | 157 SNPs associated with schizophrenia | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 8 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 3.6916 |
Section I. Genetics and epigenetics annotation
 CV:PheWAS
CV:PheWAS
| SNP ID | Chromosome | Position | Allele | P | Function | Gene | Up/Down Distance |
|---|---|---|---|---|---|---|---|
| rs242557 | 17 | 0 | null | 1.576 | MAPT | null |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs12947718 | chr17 | 43493100 | MAPT | 4137 | 0 | cis | ||
| rs17631303 | chr17 | 43516401 | MAPT | 4137 | 0 | cis | ||
| rs3946526 | chr17 | 43541655 | MAPT | 4137 | 1.764E-4 | cis | ||
| rs2696425 | chr17 | 43666905 | MAPT | 4137 | 8.37E-4 | cis | ||
| rs418891 | chr17 | 43693537 | MAPT | 4137 | 0 | cis | ||
| rs413778 | chr17 | 43716884 | MAPT | 4137 | 0 | cis | ||
| rs453997 | chr17 | 43727060 | MAPT | 4137 | 5.899E-4 | cis | ||
| rs422112 | chr17 | 43728136 | MAPT | 4137 | 0 | cis | ||
| rs241033 | chr17 | 43733982 | MAPT | 4137 | 5.899E-4 | cis | ||
| rs241032 | chr17 | 43734144 | MAPT | 4137 | 0 | cis | ||
| rs241031 | chr17 | 43734303 | MAPT | 4137 | 5.899E-4 | cis | ||
| rs17760733 | chr17 | 43746275 | MAPT | 4137 | 0.01 | cis | ||
| rs17687571 | chr17 | 43750009 | MAPT | 4137 | 0 | cis | ||
| rs17687667 | chr17 | 43754098 | MAPT | 4137 | 6.42E-4 | cis | ||
| rs17688002 | chr17 | 43762593 | MAPT | 4137 | 5.899E-4 | cis | ||
| rs17688068 | chr17 | 43763934 | MAPT | 4137 | 0.02 | cis | ||
| rs17688391 | chr17 | 43772108 | MAPT | 4137 | 9.156E-4 | cis | ||
| rs17688434 | chr17 | 43772539 | MAPT | 4137 | 0 | cis | ||
| rs12150610 | chr17 | 43775478 | MAPT | 4137 | 0 | cis | ||
| rs12150547 | chr17 | 43775545 | MAPT | 4137 | 5.899E-4 | cis | ||
| rs17762165 | chr17 | 43778601 | MAPT | 4137 | 8.734E-4 | cis | ||
| rs17688922 | chr17 | 43779350 | MAPT | 4137 | 0 | cis | ||
| rs17688944 | chr17 | 43779418 | MAPT | 4137 | 5.878E-4 | cis | ||
| rs1526128 | chr17 | 43779623 | MAPT | 4137 | 7.6E-4 | cis | ||
| rs1526129 | chr17 | 43779656 | MAPT | 4137 | 0 | cis | ||
| rs1724425 | chr17 | 43781746 | MAPT | 4137 | 0.01 | cis | ||
| rs17563501 | chr17 | 43801694 | MAPT | 4137 | 7.043E-4 | cis | ||
| rs2902662 | chr17 | 43806924 | MAPT | 4137 | 0 | cis | ||
| rs17563787 | chr17 | 43813251 | MAPT | 4137 | 5.878E-4 | cis | ||
| rs17563827 | chr17 | 43818221 | MAPT | 4137 | 7.176E-4 | cis | ||
| rs12150672 | chr17 | 43826636 | MAPT | 4137 | 0.01 | cis | ||
| rs17426106 | chr17 | 43828934 | MAPT | 4137 | 5.452E-4 | cis | ||
| rs17426195 | chr17 | 43832366 | MAPT | 4137 | 7.513E-4 | cis | ||
| rs6503447 | chr17 | 43834911 | MAPT | 4137 | 0.06 | cis | ||
| rs4074462 | chr17 | 43855227 | MAPT | 4137 | 0 | cis | ||
| rs17689471 | chr17 | 43892972 | MAPT | 4137 | 7.043E-4 | cis | ||
| rs17762769 | chr17 | 43893402 | MAPT | 4137 | 0 | cis | ||
| rs171440 | chr17 | 43893486 | MAPT | 4137 | 0.08 | cis | ||
| rs8072451 | chr17 | 43893715 | MAPT | 4137 | 0 | cis | ||
| rs4277389 | chr17 | 43895652 | MAPT | 4137 | 5.878E-4 | cis | ||
| rs17689824 | chr17 | 43904396 | MAPT | 4137 | 6.987E-4 | cis | ||
| rs17763086 | chr17 | 43905480 | MAPT | 4137 | 0.02 | cis | ||
| rs17689882 | chr17 | 43906827 | MAPT | 4137 | 5.878E-4 | cis | ||
| rs17769552 | chr17 | 43927289 | MAPT | 4137 | 9.779E-4 | cis | ||
| rs916793 | chr17 | 43954685 | MAPT | 4137 | 7.043E-4 | cis | ||
| rs17649553 | chr17 | 43994647 | MAPT | 4137 | 0 | cis | ||
| rs17649641 | chr17 | 43997371 | MAPT | 4137 | 0 | cis | ||
| rs17564223 | chr17 | 43997519 | MAPT | 4137 | 0 | cis | ||
| rs17649954 | chr17 | 44001660 | MAPT | 4137 | 5.441E-4 | cis | ||
| rs17564619 | chr17 | 44002137 | MAPT | 4137 | 9.354E-4 | cis | ||
| rs17650063 | chr17 | 44002554 | MAPT | 4137 | 7.043E-4 | cis | ||
| rs17564780 | chr17 | 44005412 | MAPT | 4137 | 7.043E-4 | cis | ||
| rs17650381 | chr17 | 44013023 | MAPT | 4137 | 0 | cis | ||
| rs12150111 | chr17 | 44013937 | MAPT | 4137 | 7.043E-4 | cis | ||
| rs2316784 | chr17 | 44021698 | MAPT | 4137 | 8.505E-4 | cis | ||
| rs4327091 | chr17 | 44021716 | MAPT | 4137 | 5.878E-4 | cis | ||
| rs242559 | chr17 | 44025887 | MAPT | 4137 | 0 | cis | ||
| rs17571718 | chr17 | 44032767 | MAPT | 4137 | 4.555E-4 | cis | ||
| rs17571739 | chr17 | 44032914 | MAPT | 4137 | 5.878E-4 | cis | ||
| rs17650872 | chr17 | 44039515 | MAPT | 4137 | 7.043E-4 | cis | ||
| rs17572169 | chr17 | 44045973 | MAPT | 4137 | 5.878E-4 | cis | ||
| rs17651213 | chr17 | 44051923 | MAPT | 4137 | 0.01 | cis | ||
| rs17572361 | chr17 | 44052008 | MAPT | 4137 | 0 | cis | ||
| rs17572851 | chr17 | 44063765 | MAPT | 4137 | 0 | cis | ||
| rs17572893 | chr17 | 44064207 | MAPT | 4137 | 5.878E-4 | cis | ||
| rs17573175 | chr17 | 44071088 | MAPT | 4137 | 8.2E-4 | cis | ||
| rs1078268 | chr17 | 44075900 | MAPT | 4137 | 0 | cis | ||
| rs8070723 | chr17 | 44081063 | MAPT | 4137 | 5.878E-4 | cis | ||
| rs17652449 | chr17 | 44088936 | MAPT | 4137 | 0 | cis | ||
| rs17652502 | chr17 | 44094470 | MAPT | 4137 | 7.043E-4 | cis | ||
| rs17574228 | chr17 | 44104508 | MAPT | 4137 | 5.878E-4 | cis | ||
| rs1076222 | chr17 | 44109768 | MAPT | 4137 | 0 | cis | ||
| rs7350980 | chr17 | 44110270 | MAPT | 4137 | 0 | cis | ||
| rs4597358 | chr17 | 44110669 | MAPT | 4137 | 5.878E-4 | cis | ||
| rs17653162 | chr17 | 44111826 | MAPT | 4137 | 3.466E-4 | cis | ||
| rs17574824 | chr17 | 44115106 | MAPT | 4137 | 0 | cis | ||
| rs12150090 | chr17 | 44115885 | MAPT | 4137 | 7.043E-4 | cis | ||
| rs7221390 | chr17 | 44116949 | MAPT | 4137 | 7.043E-4 | cis | ||
| rs17575556 | chr17 | 44135826 | MAPT | 4137 | 8.802E-4 | cis | ||
| rs17659881 | chr17 | 44157596 | MAPT | 4137 | 7.043E-4 | cis | ||
| rs17660132 | chr17 | 44165802 | MAPT | 4137 | 3.611E-4 | cis | ||
| rs17577094 | chr17 | 44187491 | MAPT | 4137 | 0 | cis | ||
| rs17660907 | chr17 | 44191084 | MAPT | 4137 | 0 | cis | ||
| rs8070942 | chr17 | 44208673 | MAPT | 4137 | 5.515E-4 | cis | ||
| rs2532316 | chr17 | 44213711 | MAPT | 4137 | 5.878E-4 | cis | ||
| rs2696600 | chr17 | 44216225 | MAPT | 4137 | 0 | cis | ||
| rs1406068 | chr17 | 44234525 | MAPT | 4137 | 5.878E-4 | cis | ||
| rs2532292 | chr17 | 44237067 | MAPT | 4137 | 7.043E-4 | cis | ||
| rs2532286 | chr17 | 44241663 | MAPT | 4137 | 7.544E-4 | cis | ||
| rs2532276 | chr17 | 44246623 | MAPT | 4137 | 0 | cis | ||
| rs2532275 | chr17 | 44246996 | MAPT | 4137 | 7.043E-4 | cis | ||
| rs2532259 | chr17 | 44253363 | MAPT | 4137 | 7.043E-4 | cis | ||
| rs2732646 | chr17 | 44254378 | MAPT | 4137 | 5.878E-4 | cis | ||
| rs2732589 | chr17 | 44266021 | MAPT | 4137 | 0 | cis | ||
| rs1918788 | chr17 | 44267616 | MAPT | 4137 | 7.043E-4 | cis | ||
| rs2732596 | chr17 | 44269545 | MAPT | 4137 | 0.01 | cis | ||
| rs2732675 | chr17 | 44280187 | MAPT | 4137 | 0 | cis | ||
| rs2732674 | chr17 | 44280696 | MAPT | 4137 | 0.01 | cis | ||
| rs2668695 | chr17 | 44292125 | MAPT | 4137 | 6.7E-4 | cis | ||
| rs2532332 | chr17 | 44347726 | MAPT | 4137 | 0.08 | cis | ||
| rs2732706 | chr17 | 44351685 | MAPT | 4137 | 0 | cis | ||
| rs2668624 | chr17 | 44352871 | MAPT | 4137 | 0.01 | cis | ||
| rs199436 | chr17 | 44789284 | MAPT | 4137 | 0.17 | cis | ||
| rs142167 | chr17 | 44795233 | MAPT | 4137 | 0.12 | cis | ||
| rs199452 | chr17 | 44801339 | MAPT | 4137 | 0.13 | cis | ||
| rs199449 | chr17 | 44808901 | MAPT | 4137 | 0.15 | cis | ||
| rs199535 | chr17 | 44822661 | MAPT | 4137 | 0.04 | cis | ||
| rs199515 | chr17 | 44856640 | MAPT | 4137 | 0.1 | cis | ||
| rs12947718 | chr17 | 43493100 | MAPT | 4137 | 0.17 | trans | ||
| rs17631303 | chr17 | 43516401 | MAPT | 4137 | 0.1 | trans | ||
| rs3946526 | chr17 | 43541655 | MAPT | 4137 | 0.02 | trans | ||
| rs2696425 | chr17 | 43666905 | MAPT | 4137 | 0.06 | trans | ||
| rs418891 | chr17 | 43693537 | MAPT | 4137 | 0.17 | trans | ||
| rs413778 | chr17 | 43716884 | MAPT | 4137 | 0.13 | trans | ||
| rs453997 | chr17 | 43727060 | MAPT | 4137 | 0.05 | trans | ||
| rs422112 | chr17 | 43728136 | MAPT | 4137 | 0.13 | trans | ||
| rs241033 | chr17 | 43733982 | MAPT | 4137 | 0.05 | trans | ||
| rs241031 | chr17 | 43734303 | MAPT | 4137 | 0.05 | trans | ||
| rs17687571 | chr17 | 43750009 | MAPT | 4137 | 0.18 | trans | ||
| rs17687667 | chr17 | 43754098 | MAPT | 4137 | 0.05 | trans | ||
| rs17688002 | chr17 | 43762593 | MAPT | 4137 | 0.05 | trans | ||
| rs17688391 | chr17 | 43772108 | MAPT | 4137 | 0.07 | trans | ||
| rs17688434 | chr17 | 43772539 | MAPT | 4137 | 0.11 | trans | ||
| rs12150610 | chr17 | 43775478 | MAPT | 4137 | 0.1 | trans | ||
| rs12150547 | chr17 | 43775545 | MAPT | 4137 | 0.05 | trans | ||
| rs17762165 | chr17 | 43778601 | MAPT | 4137 | 0.06 | trans | ||
| rs17688922 | chr17 | 43779350 | MAPT | 4137 | 0.07 | trans | ||
| rs17688944 | chr17 | 43779418 | MAPT | 4137 | 0.05 | trans | ||
| rs1526128 | chr17 | 43779623 | MAPT | 4137 | 0.06 | trans | ||
| rs1526129 | chr17 | 43779656 | MAPT | 4137 | 0.08 | trans | ||
| rs17563501 | chr17 | 43801694 | MAPT | 4137 | 0.05 | trans | ||
| rs2902662 | chr17 | 43806924 | MAPT | 4137 | 0.1 | trans | ||
| rs17563787 | chr17 | 43813251 | MAPT | 4137 | 0.05 | trans | ||
| rs17563827 | chr17 | 43818221 | MAPT | 4137 | 0.05 | trans | ||
| rs17426106 | chr17 | 43828934 | MAPT | 4137 | 0.04 | trans | ||
| rs17426195 | chr17 | 43832366 | MAPT | 4137 | 0.06 | trans | ||
| rs4074462 | chr17 | 43855227 | MAPT | 4137 | 0.09 | trans | ||
| rs17689471 | chr17 | 43892972 | MAPT | 4137 | 0.05 | trans | ||
| rs17762769 | chr17 | 43893402 | MAPT | 4137 | 0.1 | trans | ||
| rs8072451 | chr17 | 43893715 | MAPT | 4137 | 0.08 | trans | ||
| rs4277389 | chr17 | 43895652 | MAPT | 4137 | 0.05 | trans | ||
| rs17689824 | chr17 | 43904396 | MAPT | 4137 | 0.05 | trans | ||
| rs17769552 | chr17 | 43927289 | MAPT | 4137 | 0.07 | trans | ||
| snp_a-2150382 | 0 | MAPT | 4137 | 0.04 | trans | |||
| rs916793 | chr17 | 43954685 | MAPT | 4137 | 0.05 | trans | ||
| rs17649553 | chr17 | 43994647 | MAPT | 4137 | 0.09 | trans | ||
| rs17649641 | chr17 | 43997371 | MAPT | 4137 | 0.17 | trans | ||
| rs17564223 | chr17 | 43997519 | MAPT | 4137 | 0.09 | trans | ||
| rs17649954 | chr17 | 44001660 | MAPT | 4137 | 0.04 | trans | ||
| rs17564619 | chr17 | 44002137 | MAPT | 4137 | 0.07 | trans | ||
| rs17650063 | chr17 | 44002554 | MAPT | 4137 | 0.05 | trans | ||
| rs17564780 | chr17 | 44005412 | MAPT | 4137 | 0.05 | trans | ||
| rs17650381 | chr17 | 44013023 | MAPT | 4137 | 0.11 | trans | ||
| rs12150111 | chr17 | 44013937 | MAPT | 4137 | 0.05 | trans | ||
| rs2316784 | chr17 | 44021698 | MAPT | 4137 | 0.06 | trans | ||
| rs4327091 | chr17 | 44021716 | MAPT | 4137 | 0.05 | trans | ||
| rs242559 | chr17 | 44025887 | MAPT | 4137 | 0.08 | trans | ||
| rs17571718 | chr17 | 44032767 | MAPT | 4137 | 0.04 | trans | ||
| rs17571739 | chr17 | 44032914 | MAPT | 4137 | 0.05 | trans | ||
| rs17650872 | chr17 | 44039515 | MAPT | 4137 | 0.05 | trans | ||
| rs17572169 | chr17 | 44045973 | MAPT | 4137 | 0.05 | trans | ||
| rs17572361 | chr17 | 44052008 | MAPT | 4137 | 0.18 | trans | ||
| rs17572851 | chr17 | 44063765 | MAPT | 4137 | 0.1 | trans | ||
| rs17572893 | chr17 | 44064207 | MAPT | 4137 | 0.05 | trans | ||
| rs17573175 | chr17 | 44071088 | MAPT | 4137 | 0.06 | trans | ||
| rs1078268 | chr17 | 44075900 | MAPT | 4137 | 0.09 | trans | ||
| rs8070723 | chr17 | 44081063 | MAPT | 4137 | 0.05 | trans | ||
| rs17652449 | chr17 | 44088936 | MAPT | 4137 | 0.15 | trans | ||
| rs17652502 | chr17 | 44094470 | MAPT | 4137 | 0.05 | trans | ||
| rs17574228 | chr17 | 44104508 | MAPT | 4137 | 0.05 | trans | ||
| rs1076222 | chr17 | 44109768 | MAPT | 4137 | 0.09 | trans | ||
| rs7350980 | chr17 | 44110270 | MAPT | 4137 | 0.08 | trans | ||
| rs4597358 | chr17 | 44110669 | MAPT | 4137 | 0.05 | trans | ||
| rs17653162 | chr17 | 44111826 | MAPT | 4137 | 0.03 | trans | ||
| rs17574824 | chr17 | 44115106 | MAPT | 4137 | 0.2 | trans | ||
| rs12150090 | chr17 | 44115885 | MAPT | 4137 | 0.05 | trans | ||
| snp_a-1855473 | 0 | MAPT | 4137 | 0.05 | trans | |||
| rs7221390 | chr17 | 44116949 | MAPT | 4137 | 0.05 | trans | ||
| snp_a-1823652 | 0 | MAPT | 4137 | 0.18 | trans | |||
| rs17575556 | chr17 | 44135826 | MAPT | 4137 | 0.06 | trans | ||
| rs17659881 | chr17 | 44157596 | MAPT | 4137 | 0.05 | trans | ||
| rs17660132 | chr17 | 44165802 | MAPT | 4137 | 0.03 | trans | ||
| rs17577094 | chr17 | 44187491 | MAPT | 4137 | 0.15 | trans | ||
| rs17660907 | chr17 | 44191084 | MAPT | 4137 | 0.07 | trans | ||
| rs8070942 | chr17 | 44208673 | MAPT | 4137 | 0.04 | trans | ||
| rs2532316 | chr17 | 44213711 | MAPT | 4137 | 0.05 | trans | ||
| rs2696600 | chr17 | 44216225 | MAPT | 4137 | 0.13 | trans | ||
| rs1406068 | chr17 | 44234525 | MAPT | 4137 | 0.05 | trans | ||
| rs2532292 | chr17 | 44237067 | MAPT | 4137 | 0.05 | trans | ||
| rs2532286 | chr17 | 44241663 | MAPT | 4137 | 0.06 | trans | ||
| rs2532276 | chr17 | 44246623 | MAPT | 4137 | 0.07 | trans | ||
| rs2532275 | chr17 | 44246996 | MAPT | 4137 | 0.05 | trans | ||
| rs2532259 | chr17 | 44253363 | MAPT | 4137 | 0.05 | trans | ||
| rs2732646 | chr17 | 44254378 | MAPT | 4137 | 0.05 | trans | ||
| rs2732589 | chr17 | 44266021 | MAPT | 4137 | 0.16 | trans | ||
| rs1918788 | chr17 | 44267616 | MAPT | 4137 | 0.05 | trans | ||
| rs2732675 | chr17 | 44280187 | MAPT | 4137 | 0.14 | trans | ||
| rs2668695 | chr17 | 44292125 | MAPT | 4137 | 0.05 | trans | ||
| rs2732706 | chr17 | 44351685 | MAPT | 4137 | 0.11 | trans | ||
| rs2668622 | 0 | MAPT | 4137 | 0.04 | trans |
Section II. Transcriptome annotation
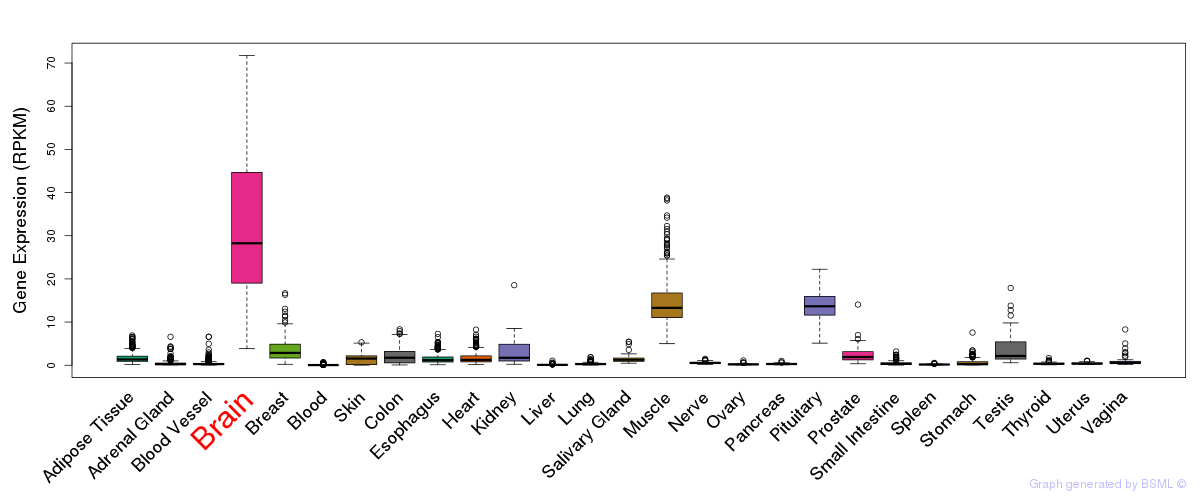
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TNS1 | 0.83 | 0.86 |
| TLN1 | 0.80 | 0.81 |
| RRBP1 | 0.79 | 0.83 |
| SLC20A2 | 0.78 | 0.82 |
| GSN | 0.77 | 0.78 |
| ANGPTL2 | 0.77 | 0.79 |
| TJP2 | 0.77 | 0.82 |
| UNC5B | 0.77 | 0.80 |
| MYLK | 0.77 | 0.79 |
| ERBB3 | 0.77 | 0.77 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NR2C2AP | -0.53 | -0.56 |
| RPL27 | -0.52 | -0.67 |
| RPL31 | -0.52 | -0.66 |
| POLB | -0.52 | -0.58 |
| RPS23 | -0.52 | -0.66 |
| FRG1 | -0.52 | -0.61 |
| RPL24 | -0.52 | -0.68 |
| CYP2R1 | -0.52 | -0.57 |
| RPS18 | -0.52 | -0.64 |
| RPL32 | -0.51 | -0.64 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005200 | structural constituent of cytoskeleton | TAS | 2498079 | |
| GO:0008034 | lipoprotein binding | IPI | 7972031 | |
| GO:0017124 | SH3 domain binding | IPI | 9763511 | |
| GO:0008017 | microtubule binding | IDA | 1918161 | |
| GO:0008017 | microtubule binding | ISS | - | |
| GO:0019899 | enzyme binding | IPI | 9736630 |12888622 | |
| GO:0034187 | apolipoprotein E binding | IPI | 7566652 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0048699 | generation of neurons | NAS | neuron, neurogenesis (GO term level: 7) | 8522593 |
| GO:0045773 | positive regulation of axon extension | IDA | axon (GO term level: 15) | 1389180 |
| GO:0045773 | positive regulation of axon extension | ISS | axon (GO term level: 15) | - |
| GO:0000226 | microtubule cytoskeleton organization | IDA | 1057175 | |
| GO:0000226 | microtubule cytoskeleton organization | ISS | - | |
| GO:0007026 | negative regulation of microtubule depolymerization | IEA | - | |
| GO:0031116 | positive regulation of microtubule polymerization | IDA | 1421571 | |
| GO:0031116 | positive regulation of microtubule polymerization | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0042995 | cell projection | IEA | axon (GO term level: 4) | - |
| GO:0030426 | growth cone | IDA | axon, dendrite (GO term level: 5) | 8642405 |
| GO:0030426 | growth cone | ISS | axon, dendrite (GO term level: 5) | - |
| GO:0030424 | axon | IDA | neuron, axon, Neurotransmitter (GO term level: 6) | 8642405 |
| GO:0030424 | axon | ISS | neuron, axon, Neurotransmitter (GO term level: 6) | - |
| GO:0005829 | cytosol | TAS | 10747907 | |
| GO:0005856 | cytoskeleton | IEA | - | |
| GO:0005875 | microtubule associated complex | TAS | 10747907 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005886 | plasma membrane | IDA | 8522593 | |
| GO:0005886 | plasma membrane | ISS | - | |
| GO:0045298 | tubulin complex | IDA | 8642405 | |
| GO:0045298 | tubulin complex | ISS | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AATF | CHE-1 | CHE1 | DED | apoptosis antagonizing transcription factor | - | HPRD,BioGRID | 14697667 |
| APOE | AD2 | LPG | MGC1571 | apoprotein | apolipoprotein E | Reconstituted Complex | BioGRID | 8620924 |
| APOE | AD2 | LPG | MGC1571 | apoprotein | apolipoprotein E | - | HPRD | 7566652 |9147408 |
| CALM1 | CALML2 | CAMI | DD132 | PHKD | calmodulin 1 (phosphorylase kinase, delta) | - | HPRD,BioGRID | 2123288 |
| FYN | MGC45350 | SLK | SYN | FYN oncogene related to SRC, FGR, YES | - | HPRD,BioGRID | 11826099 |
| GSK3B | - | glycogen synthase kinase 3 beta | GSK3beta phosphorylates Ftau.This interaction was modelled on a demonstrated interaction between rabbit GSK3beta and human Ftau. | BIND | 14636947 |
| MARK4 | FLJ90097 | KIAA1860 | MARKL1 | Nbla00650 | MAP/microtubule affinity-regulating kinase 4 | - | HPRD,BioGRID | 14594945 |
| OGT | FLJ23071 | HRNT1 | MGC22921 | O-GLCNAC | O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase) | - | HPRD,BioGRID | 8910513 |12911634 |
| PIN1 | DOD | UBL5 | peptidylprolyl cis/trans isomerase, NIMA-interacting 1 | - | HPRD,BioGRID | 10391244 |
| PSEN1 | AD3 | FAD | PS1 | S182 | presenilin 1 | - | HPRD,BioGRID | 9689133 |
| S100B | NEF | S100 | S100beta | S100 calcium binding protein B | - | HPRD | 11264299 |
| S100B | NEF | S100 | S100beta | S100 calcium binding protein B | - | HPRD,BioGRID | 2833519 |
| SNCA | MGC110988 | NACP | PARK1 | PARK4 | PD1 | synuclein, alpha (non A4 component of amyloid precursor) | - | HPRD,BioGRID | 10464279 |
| SPTB | HSpTB1 | spectrin, beta, erythrocytic | - | HPRD,BioGRID | 6743699 |
| STXBP1 | EIEE4 | MUNC18-1 | UNC18 | hUNC18 | rbSec1 | syntaxin binding protein 1 | - | HPRD,BioGRID | 12963086 |
| TUBB2A | TUBB | TUBB2 | dJ40E16.7 | tubulin, beta 2A | - | HPRD | 9922135 |
| USP9Y | AZF | AZF1 | AZFA | DFFRY | FLJ33043 | SP3 | ubiquitin specific peptidase 9, Y-linked (fat facets-like, Drosophila) | - | HPRD,BioGRID | 1723470 |
| YWHAA | - | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, alpha polypeptide | Affinity Capture-Western | BioGRID | 12176984 |
| YWHAB | GW128 | HS1 | KCIP-1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | - | HPRD,BioGRID | 10840038 |
| YWHAZ | KCIP-1 | MGC111427 | MGC126532 | MGC138156 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | Tau interacts with 14-3-3-zeta. | BIND | 12176984 |
| YWHAZ | KCIP-1 | MGC111427 | MGC126532 | MGC138156 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | - | HPRD,BioGRID | 10840038 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG ALZHEIMERS DISEASE | 169 | 110 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BIOPEPTIDES PATHWAY | 45 | 35 | All SZGR 2.0 genes in this pathway |
| BIOCARTA P35ALZHEIMERS PATHWAY | 11 | 9 | All SZGR 2.0 genes in this pathway |
| PID LYSOPHOSPHOLIPID PATHWAY | 66 | 53 | All SZGR 2.0 genes in this pathway |
| PID REELIN PATHWAY | 29 | 29 | All SZGR 2.0 genes in this pathway |
| PID LKB1 PATHWAY | 47 | 37 | All SZGR 2.0 genes in this pathway |
| PID P38 GAMMA DELTA PATHWAY | 11 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | 40 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | 13 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOSIS | 148 | 94 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOTIC EXECUTION PHASE | 54 | 37 | All SZGR 2.0 genes in this pathway |
| LIU SOX4 TARGETS DN | 309 | 191 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE DN | 244 | 147 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL UP | 380 | 215 | All SZGR 2.0 genes in this pathway |
| DOANE RESPONSE TO ANDROGEN UP | 184 | 125 | All SZGR 2.0 genes in this pathway |
| DOANE BREAST CANCER ESR1 UP | 112 | 72 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER 20Q13 AMPLIFICATION UP | 119 | 66 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS G UP | 238 | 135 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS BASAL | 330 | 217 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE UP | 134 | 93 | All SZGR 2.0 genes in this pathway |
| TERAMOTO OPN TARGETS CLUSTER 8 | 8 | 7 | All SZGR 2.0 genes in this pathway |
| YANG BREAST CANCER ESR1 UP | 36 | 22 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| ASTON MAJOR DEPRESSIVE DISORDER DN | 160 | 110 | All SZGR 2.0 genes in this pathway |
| FRASOR RESPONSE TO ESTRADIOL UP | 37 | 28 | All SZGR 2.0 genes in this pathway |
| ZHANG TARGETS OF EWSR1 FLI1 FUSION | 88 | 68 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR UP | 240 | 152 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN DN | 153 | 120 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL LONG TERM | 302 | 191 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS DN | 31 | 23 | All SZGR 2.0 genes in this pathway |
| KAYO CALORIE RESTRICTION MUSCLE UP | 95 | 64 | All SZGR 2.0 genes in this pathway |
| MCDOWELL ACUTE LUNG INJURY DN | 48 | 33 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 4 | 307 | 185 | All SZGR 2.0 genes in this pathway |
| KONDO PROSTATE CANCER HCP WITH H3K27ME3 | 97 | 72 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| LEE METASTASIS AND ALTERNATIVE SPLICING DN | 45 | 31 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE DN | 258 | 160 | All SZGR 2.0 genes in this pathway |
| MASSARWEH RESPONSE TO ESTRADIOL | 61 | 47 | All SZGR 2.0 genes in this pathway |
| OUILLETTE CLL 13Q14 DELETION DN | 60 | 38 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BRAIN DN | 85 | 58 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B UP | 172 | 109 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| TOYOTA TARGETS OF MIR34B AND MIR34C | 463 | 262 | All SZGR 2.0 genes in this pathway |
| ZHENG GLIOBLASTOMA PLASTICITY DN | 58 | 39 | All SZGR 2.0 genes in this pathway |
| HUANG DASATINIB RESISTANCE DN | 69 | 44 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| HOFFMANN PRE BI TO LARGE PRE BII LYMPHOCYTE UP | 36 | 24 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE UP | 442 | 263 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER DN | 134 | 83 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA PRONEURAL | 177 | 132 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 6HR UP | 229 | 149 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS VIA AKT1 UP | 281 | 183 | All SZGR 2.0 genes in this pathway |
| FARDIN HYPOXIA 11 | 32 | 29 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION UP | 271 | 165 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM A | 182 | 108 | All SZGR 2.0 genes in this pathway |
| GHANDHI BYSTANDER IRRADIATION DN | 12 | 10 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-181 | 4068 | 4074 | m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-214 | 2039 | 2045 | m8 | hsa-miR-214brain | ACAGCAGGCACAGACAGGCAG |
| miR-219 | 3175 | 3181 | m8 | hsa-miR-219brain | UGAUUGUCCAAACGCAAUUCU |
| miR-27 | 4065 | 4071 | 1A | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-34/449 | 1187 | 1193 | m8 | hsa-miR-34abrain | UGGCAGUGUCUUAGCUGGUUGUU |
| hsa-miR-34c | AGGCAGUGUAGUUAGCUGAUUGC | ||||
| hsa-miR-449 | UGGCAGUGUAUUGUUAGCUGGU | ||||
| hsa-miR-449b | AGGCAGUGUAUUGUUAGCUGGC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.