Gene Page: MTRR
Summary ?
| GeneID | 4552 |
| Symbol | MTRR |
| Synonyms | MSR|cblE |
| Description | 5-methyltetrahydrofolate-homocysteine methyltransferase reductase |
| Reference | MIM:602568|HGNC:HGNC:7473|Ensembl:ENSG00000124275|HPRD:03979|Vega:OTTHUMG00000090477 |
| Gene type | protein-coding |
| Map location | 5p15.31 |
| Pascal p-value | 0.157 |
| Sherlock p-value | 0.539 |
| Fetal beta | -0.646 |
| eGene | Caudate basal ganglia Cortex Frontal Cortex BA9 Nucleus accumbens basal ganglia Putamen basal ganglia Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs327588 | chr5 | 7908358 | MTRR | 4552 | 0.04 | cis | ||
| rs17243407 | 5 | 7858121 | MTRR | ENSG00000124275.10 | 1.732E-7 | 0 | 1221 | gtex_brain_putamen_basal |
| rs7716373 | 5 | 7860908 | MTRR | ENSG00000124275.10 | 3.182E-7 | 0 | 4008 | gtex_brain_putamen_basal |
| rs13154459 | 5 | 7861619 | MTRR | ENSG00000124275.10 | 1.524E-7 | 0 | 4719 | gtex_brain_putamen_basal |
| rs17184211 | 5 | 7866106 | MTRR | ENSG00000124275.10 | 3.556E-7 | 0 | 9206 | gtex_brain_putamen_basal |
| rs1801394 | 5 | 7870973 | MTRR | ENSG00000124275.10 | 8.361E-10 | 0 | 14073 | gtex_brain_putamen_basal |
| rs13181011 | 5 | 7874424 | MTRR | ENSG00000124275.10 | 3.509E-7 | 0 | 17524 | gtex_brain_putamen_basal |
| rs11949523 | 5 | 7881027 | MTRR | ENSG00000124275.10 | 1.74E-7 | 0 | 24127 | gtex_brain_putamen_basal |
| rs62342527 | 5 | 7882211 | MTRR | ENSG00000124275.10 | 1.556E-7 | 0 | 25311 | gtex_brain_putamen_basal |
| rs35035568 | 5 | 7886585 | MTRR | ENSG00000124275.10 | 1.831E-7 | 0 | 29685 | gtex_brain_putamen_basal |
| rs13189728 | 5 | 7887284 | MTRR | ENSG00000124275.10 | 1.281E-7 | 0 | 30384 | gtex_brain_putamen_basal |
| rs13190084 | 5 | 7887488 | MTRR | ENSG00000124275.10 | 1.278E-7 | 0 | 30588 | gtex_brain_putamen_basal |
| rs144069526 | 5 | 7887847 | MTRR | ENSG00000124275.10 | 1.271E-7 | 0 | 30947 | gtex_brain_putamen_basal |
| rs62342530 | 5 | 7888198 | MTRR | ENSG00000124275.10 | 1.261E-7 | 0 | 31298 | gtex_brain_putamen_basal |
| rs17267737 | 5 | 7893808 | MTRR | ENSG00000124275.10 | 1.137E-7 | 0 | 36908 | gtex_brain_putamen_basal |
| rs34966478 | 5 | 7896606 | MTRR | ENSG00000124275.10 | 2.296E-8 | 0 | 39706 | gtex_brain_putamen_basal |
| rs9282787 | 5 | 7900598 | MTRR | ENSG00000124275.10 | 2.141E-7 | 0 | 43698 | gtex_brain_putamen_basal |
| rs12108985 | 5 | 7907309 | MTRR | ENSG00000124275.10 | 2.141E-7 | 0 | 50409 | gtex_brain_putamen_basal |
| rs17245023 | 5 | 7909855 | MTRR | ENSG00000124275.10 | 9.72E-8 | 0 | 52955 | gtex_brain_putamen_basal |
| rs2128266 | 5 | 7911470 | MTRR | ENSG00000124275.10 | 2.141E-7 | 0 | 54570 | gtex_brain_putamen_basal |
| rs13173919 | 5 | 7919212 | MTRR | ENSG00000124275.10 | 4.759E-8 | 0 | 62312 | gtex_brain_putamen_basal |
| rs13178414 | 5 | 7919602 | MTRR | ENSG00000124275.10 | 4.495E-8 | 0 | 62702 | gtex_brain_putamen_basal |
| rs10462646 | 5 | 7920322 | MTRR | ENSG00000124275.10 | 1.419E-8 | 0 | 63422 | gtex_brain_putamen_basal |
| rs13170757 | 5 | 7922542 | MTRR | ENSG00000124275.10 | 1.332E-7 | 0 | 65642 | gtex_brain_putamen_basal |
| rs16879466 | 5 | 7930195 | MTRR | ENSG00000124275.10 | 3.206E-8 | 0 | 73295 | gtex_brain_putamen_basal |
| rs10440693 | 5 | 7930866 | MTRR | ENSG00000124275.10 | 3.325E-8 | 0 | 73966 | gtex_brain_putamen_basal |
| rs999610 | 5 | 7931390 | MTRR | ENSG00000124275.10 | 9.72E-8 | 0 | 74490 | gtex_brain_putamen_basal |
| rs1968991 | 5 | 7932199 | MTRR | ENSG00000124275.10 | 9.72E-8 | 0 | 75299 | gtex_brain_putamen_basal |
| rs1493447 | 5 | 7935039 | MTRR | ENSG00000124275.10 | 9.72E-8 | 0 | 78139 | gtex_brain_putamen_basal |
| rs34520786 | 5 | 7937555 | MTRR | ENSG00000124275.10 | 8.592E-9 | 0 | 80655 | gtex_brain_putamen_basal |
| rs1493446 | 5 | 7938528 | MTRR | ENSG00000124275.10 | 1.556E-7 | 0 | 81628 | gtex_brain_putamen_basal |
| rs62358784 | 5 | 7946742 | MTRR | ENSG00000124275.10 | 2.237E-6 | 0 | 89842 | gtex_brain_putamen_basal |
| rs767084 | 5 | 7947648 | MTRR | ENSG00000124275.10 | 2.237E-6 | 0 | 90748 | gtex_brain_putamen_basal |
| rs78787612 | 5 | 7948112 | MTRR | ENSG00000124275.10 | 2.234E-6 | 0 | 91212 | gtex_brain_putamen_basal |
| rs1542781 | 5 | 7952536 | MTRR | ENSG00000124275.10 | 2.272E-6 | 0 | 95636 | gtex_brain_putamen_basal |
| rs13157729 | 5 | 7953623 | MTRR | ENSG00000124275.10 | 3.11E-7 | 0 | 96723 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
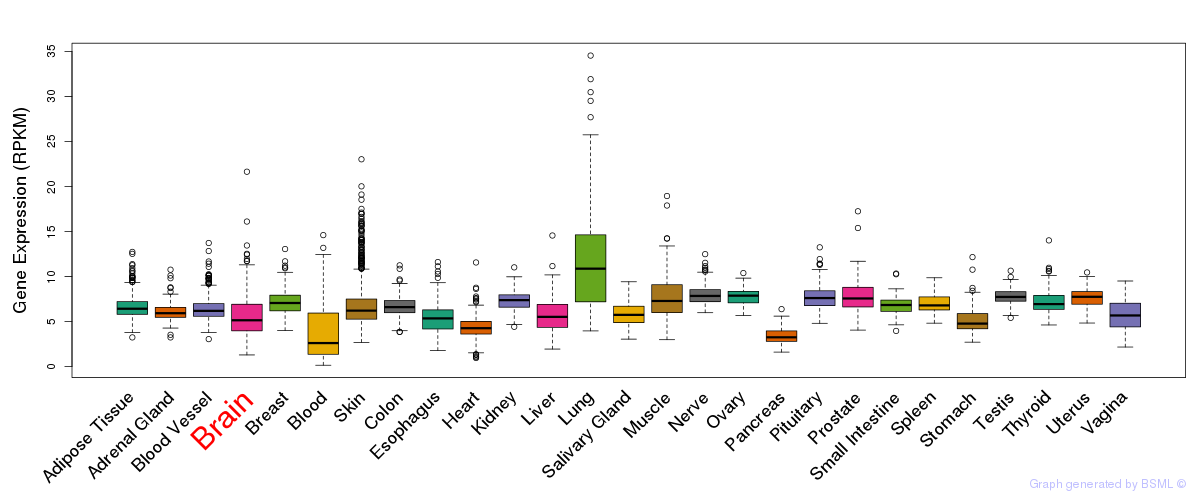
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005506 | iron ion binding | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0008168 | methyltransferase activity | IEA | - | |
| GO:0010181 | FMN binding | IEA | - | |
| GO:0010181 | FMN binding | TAS | 9501215 | |
| GO:0009055 | electron carrier activity | IEA | - | |
| GO:0016491 | oxidoreductase activity | IEA | - | |
| GO:0030586 | [methionine synthase] reductase activity | TAS | 9501215 | |
| GO:0050660 | FAD binding | TAS | 9501215 | |
| GO:0050661 | NADP binding | TAS | 9501215 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0009086 | methionine biosynthetic process | IEA | - | |
| GO:0008652 | amino acid biosynthetic process | IEA | - | |
| GO:0055114 | oxidation reduction | TAS | 9501215 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | TAS | 9501215 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005739 | mitochondrion | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| TIEN INTESTINE PROBIOTICS 6HR DN | 167 | 100 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| MIDORIKAWA AMPLIFIED IN LIVER CANCER | 55 | 38 | All SZGR 2.0 genes in this pathway |
| LOCKWOOD AMPLIFIED IN LUNG CANCER | 214 | 139 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP | 262 | 186 | All SZGR 2.0 genes in this pathway |
| BACOLOD RESISTANCE TO ALKYLATING AGENTS DN | 60 | 45 | All SZGR 2.0 genes in this pathway |
| LEIN OLIGODENDROCYTE MARKERS | 74 | 53 | All SZGR 2.0 genes in this pathway |
| WONG MITOCHONDRIA GENE MODULE | 217 | 122 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS DN | 414 | 237 | All SZGR 2.0 genes in this pathway |
| ACOSTA PROLIFERATION INDEPENDENT MYC TARGETS UP | 84 | 51 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| ZWANG EGF PERSISTENTLY DN | 61 | 36 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-361 | 538 | 545 | 1A,m8 | hsa-miR-361brain | UUAUCAGAAUCUCCAGGGGUAC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.