Gene Page: ATP1A1
Summary ?
| GeneID | 476 |
| Symbol | ATP1A1 |
| Synonyms | - |
| Description | ATPase Na+/K+ transporting subunit alpha 1 |
| Reference | MIM:182310|HGNC:HGNC:799|Ensembl:ENSG00000163399|HPRD:01662|Vega:OTTHUMG00000012109 |
| Gene type | protein-coding |
| Map location | 1p21 |
| Pascal p-value | 0.251 |
| Sherlock p-value | 0.966 |
| Fetal beta | -2.189 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | ION BALANCE G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_clathrin G2Cdb.human_mGluR5 G2Cdb.human_Synaptosome G2Cdb.humanARC G2Cdb.humanNRC G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Darnell FMRP targets Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0235 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00814 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg20309677 | 1 | 116925753 | ATP1A1 | 4.477E-4 | 0.552 | 0.045 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs12123144 | chr1 | 76550208 | ATP1A1 | 476 | 0.17 | trans |
Section II. Transcriptome annotation
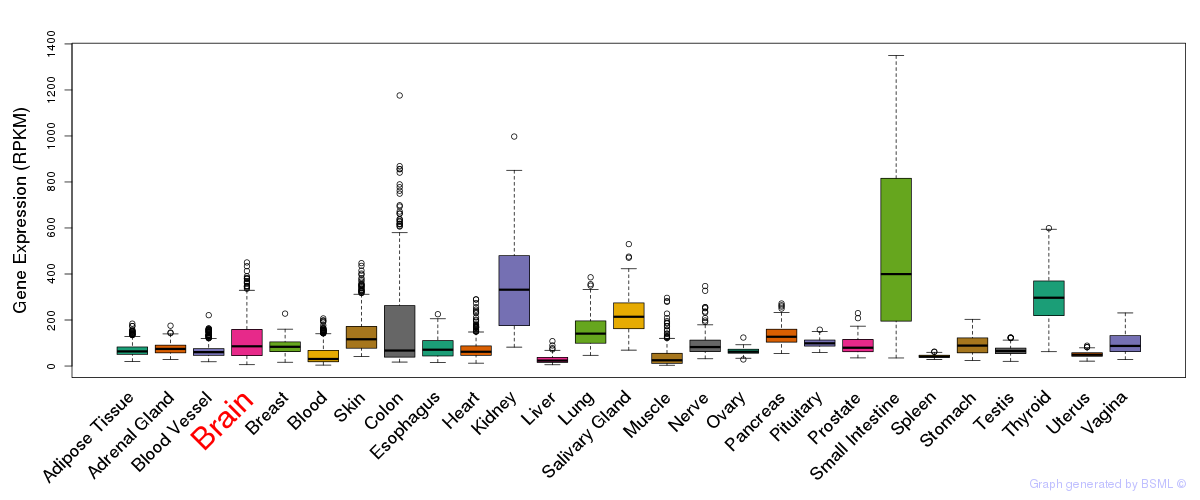
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PPL | 0.66 | 0.59 |
| AC110760.2 | 0.59 | 0.60 |
| ESRRG | 0.57 | 0.59 |
| ECM1 | 0.57 | 0.48 |
| FBLN7 | 0.56 | 0.56 |
| GALR1 | 0.56 | 0.45 |
| VSTM2A | 0.54 | 0.65 |
| RET | 0.54 | 0.62 |
| PKP1 | 0.54 | 0.40 |
| KCNS2 | 0.54 | 0.58 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| IMPA2 | -0.24 | -0.36 |
| AC098691.2 | -0.22 | -0.29 |
| SIGIRR | -0.22 | -0.33 |
| RPL37 | -0.21 | -0.28 |
| TAX1BP3 | -0.21 | -0.21 |
| UXT | -0.21 | -0.19 |
| ANP32C | -0.21 | -0.25 |
| EMX2 | -0.21 | -0.32 |
| EIF4EBP1 | -0.21 | -0.18 |
| MPST | -0.20 | -0.31 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0000287 | magnesium ion binding | IEA | - | |
| GO:0003869 | 4-nitrophenylphosphatase activity | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 11027149 |15671290 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0005391 | sodium:potassium-exchanging ATPase activity | IEA | - | |
| GO:0005391 | sodium:potassium-exchanging ATPase activity | TAS | 1975705 | |
| GO:0016787 | hydrolase activity | IEA | - | |
| GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances | IEA | - | |
| GO:0015662 | ATPase activity, coupled to transmembrane movement of ions, phosphorylative mechanism | IEA | - | |
| GO:0015077 | monovalent inorganic cation transmembrane transporter activity | IEA | - | |
| GO:0030955 | potassium ion binding | IEA | - | |
| GO:0031402 | sodium ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0002026 | regulation of the force of heart contraction | IEA | - | |
| GO:0008217 | regulation of blood pressure | IEA | - | |
| GO:0008152 | metabolic process | IEA | - | |
| GO:0006814 | sodium ion transport | IEA | - | |
| GO:0006812 | cation transport | IEA | - | |
| GO:0006813 | potassium ion transport | IEA | - | |
| GO:0006810 | transport | IEA | - | |
| GO:0042493 | response to drug | IEA | - | |
| GO:0030317 | sperm motility | ISS | - | |
| GO:0030641 | regulation of cellular pH | ISS | - | |
| GO:0031947 | negative regulation of glucocorticoid biosynthetic process | IEA | - | |
| GO:0045822 | negative regulation of heart contraction | IEA | - | |
| GO:0045823 | positive regulation of heart contraction | IEA | - | |
| GO:0045989 | positive regulation of striated muscle contraction | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005792 | microsome | IEA | - | |
| GO:0005624 | membrane fraction | TAS | 1975705 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | ISS | - | |
| GO:0005890 | sodium:potassium-exchanging ATPase complex | TAS | 1975705 | |
| GO:0016323 | basolateral plasma membrane | IEA | - | |
| GO:0042383 | sarcolemma | IEA | - | |
| GO:0042470 | melanosome | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ADD1 | ADDA | MGC3339 | MGC44427 | adducin 1 (alpha) | - | HPRD | 10516168 |
| CFL1 | CFL | cofilin 1 (non-muscle) | - | HPRD | 11139403 |
| EZH2 | ENX-1 | EZH1 | KMT6 | MGC9169 | enhancer of zeste homolog 2 (Drosophila) | Two-hybrid | BioGRID | 16169070 |
| FXYD1 | MGC44983 | PLM | FXYD domain containing ion transport regulator 1 | - | HPRD,BioGRID | 14597563 |
| FXYD7 | FLJ25096 | FXYD domain containing ion transport regulator 7 | - | HPRD | 12093728 |
| MAPK1 | ERK | ERK2 | ERT1 | MAPK2 | P42MAPK | PRKM1 | PRKM2 | p38 | p40 | p41 | p41mapk | mitogen-activated protein kinase 1 | Biochemical Activity | BioGRID | 15069082 |
| MAPK3 | ERK1 | HS44KDAP | HUMKER1A | MGC20180 | P44ERK1 | P44MAPK | PRKM3 | mitogen-activated protein kinase 3 | - | HPRD,BioGRID | 15069082 |
| PRKCA | AAG6 | MGC129900 | MGC129901 | PKC-alpha | PKCA | PRKACA | protein kinase C, alpha | Biochemical Activity | BioGRID | 15069082 |
| TNFRSF1A | CD120a | FPF | MGC19588 | TBP1 | TNF-R | TNF-R-I | TNF-R55 | TNFAR | TNFR1 | TNFR55 | TNFR60 | p55 | p55-R | p60 | tumor necrosis factor receptor superfamily, member 1A | - | HPRD | 14743216 |
| TNFRSF1B | CD120b | TBPII | TNF-R-II | TNF-R75 | TNFBR | TNFR1B | TNFR2 | TNFR80 | p75 | p75TNFR | tumor necrosis factor receptor superfamily, member 1B | - | HPRD | 14743216 |
| TPT1 | FLJ27337 | HRF | TCTP | p02 | tumor protein, translationally-controlled 1 | - | HPRD | 15383549 |
| TRADD | Hs.89862 | MGC11078 | TNFRSF1A-associated via death domain | - | HPRD | 14743216 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CARDIAC MUSCLE CONTRACTION | 80 | 51 | All SZGR 2.0 genes in this pathway |
| KEGG ALDOSTERONE REGULATED SODIUM REABSORPTION | 42 | 29 | All SZGR 2.0 genes in this pathway |
| KEGG PROXIMAL TUBULE BICARBONATE RECLAMATION | 23 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | 413 | 270 | All SZGR 2.0 genes in this pathway |
| REACTOME ION TRANSPORT BY P TYPE ATPASES | 34 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME ION CHANNEL TRANSPORT | 55 | 42 | All SZGR 2.0 genes in this pathway |
| WATANABE RECTAL CANCER RADIOTHERAPY RESPONSIVE UP | 108 | 67 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN | 455 | 304 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER BASAL VS MESENCHYMAL UP | 121 | 45 | All SZGR 2.0 genes in this pathway |
| DITTMER PTHLH TARGETS UP | 112 | 68 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| CHEBOTAEV GR TARGETS UP | 77 | 62 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK UP | 271 | 175 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL TGFB1 TARGETS UP | 169 | 127 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| ALCALA APOPTOSIS | 88 | 60 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 12HR UP | 116 | 79 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP | 487 | 286 | All SZGR 2.0 genes in this pathway |
| MORI SMALL PRE BII LYMPHOCYTE DN | 76 | 52 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO DN | 200 | 112 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB TARGETS | 74 | 41 | All SZGR 2.0 genes in this pathway |
| SHEPARD CRUSH AND BURN MUTANT DN | 185 | 111 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO UP | 205 | 126 | All SZGR 2.0 genes in this pathway |
| JECHLINGER EPITHELIAL TO MESENCHYMAL TRANSITION DN | 66 | 47 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| ZHANG TARGETS OF EWSR1 FLI1 FUSION | 88 | 68 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND UP | 77 | 52 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 1 UP | 182 | 119 | All SZGR 2.0 genes in this pathway |
| XU CREBBP TARGETS DN | 44 | 31 | All SZGR 2.0 genes in this pathway |
| DELASERNA MYOD TARGETS UP | 89 | 51 | All SZGR 2.0 genes in this pathway |
| ZAMORA NOS2 TARGETS DN | 96 | 71 | All SZGR 2.0 genes in this pathway |
| JIANG AGING CEREBRAL CORTEX DN | 54 | 43 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE UP | 393 | 244 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS UP | 170 | 107 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR UP | 294 | 199 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC | 420 | 269 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS PROLIFERATION UP | 178 | 108 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS UNANNOTATED DN | 193 | 112 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 7 | 76 | 46 | All SZGR 2.0 genes in this pathway |
| PANGAS TUMOR SUPPRESSION BY SMAD1 AND SMAD5 DN | 157 | 106 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS UP | 259 | 159 | All SZGR 2.0 genes in this pathway |
| BILANGES SERUM SENSITIVE VIA TSC2 | 39 | 25 | All SZGR 2.0 genes in this pathway |
| HOLLEMAN DAUNORUBICIN B ALL UP | 10 | 9 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL UP | 289 | 184 | All SZGR 2.0 genes in this pathway |