Gene Page: PALM
Summary ?
| GeneID | 5064 |
| Symbol | PALM |
| Synonyms | - |
| Description | paralemmin |
| Reference | MIM:608134|HGNC:HGNC:8594|Ensembl:ENSG00000099864|HPRD:09734|Vega:OTTHUMG00000181785 |
| Gene type | protein-coding |
| Map location | 19p13.3 |
| Pascal p-value | 0.16 |
| Sherlock p-value | 0.611 |
| Fetal beta | 0.518 |
| DMG | 1 (# studies) |
| eGene | Nucleus accumbens basal ganglia Myers' cis & trans |
| Support | STRUCTURAL PLASTICITY G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg01729837 | 19 | 719622 | PALM | 7.48E-5 | 0.383 | 0.025 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs5995385 | chr22 | 37519863 | PALM | 5064 | 0.2 | trans |
Section II. Transcriptome annotation
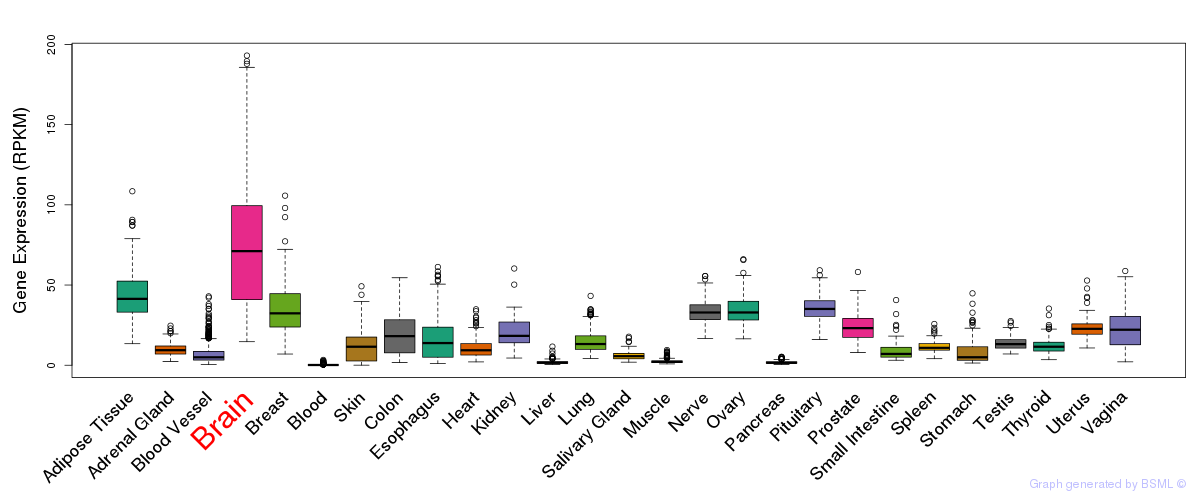
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TMX2 | 0.80 | 0.81 |
| PEX19 | 0.78 | 0.83 |
| PRDX3 | 0.77 | 0.76 |
| PKM2 | 0.74 | 0.79 |
| PPT1 | 0.73 | 0.78 |
| SDHD | 0.73 | 0.78 |
| PSAP | 0.72 | 0.75 |
| DAZAP2 | 0.71 | 0.73 |
| TM7SF3 | 0.71 | 0.73 |
| ATG7 | 0.71 | 0.79 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AC005921.3 | -0.53 | -0.57 |
| ZNF814 | -0.50 | -0.53 |
| AC010300.1 | -0.46 | -0.46 |
| IL3RA | -0.45 | -0.52 |
| AC004148.1 | -0.45 | -0.49 |
| C21orf32 | -0.45 | -0.47 |
| ANKRD36 | -0.45 | -0.49 |
| ANKRD36B | -0.43 | -0.48 |
| ARHGAP8 | -0.43 | -0.43 |
| AC109829.1 | -0.43 | -0.38 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0031750 | D3 dopamine receptor binding | IEA | dopamine (GO term level: 7) | - |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007194 | negative regulation of adenylate cyclase activity | IEA | - | |
| GO:0007010 | cytoskeleton organization | IEA | - | |
| GO:0008360 | regulation of cell shape | IEA | - | |
| GO:0008104 | protein localization | IEA | - | |
| GO:0006928 | cell motion | TAS | 9813098 | |
| GO:0030818 | negative regulation of cAMP biosynthetic process | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0016023 | cytoplasmic membrane-bounded vesicle | TAS | 9813098 | |
| GO:0005886 | plasma membrane | NAS | 9615234 | |
| GO:0005887 | integral to plasma membrane | TAS | 9813098 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION ERYTHROCYTE UP | 157 | 104 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS UP | 149 | 84 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| ROSS AML WITH AML1 ETO FUSION | 76 | 55 | All SZGR 2.0 genes in this pathway |
| HOEGERKORP CD44 TARGETS DIRECT DN | 14 | 7 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED DN | 246 | 180 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY NO BLOOD DN | 150 | 93 | All SZGR 2.0 genes in this pathway |
| MCCLUNG COCAINE REWARD 5D | 79 | 62 | All SZGR 2.0 genes in this pathway |
| NIELSEN LIPOSARCOMA UP | 18 | 13 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY DN | 145 | 88 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| AMBROSINI FLAVOPIRIDOL TREATMENT TP53 | 109 | 63 | All SZGR 2.0 genes in this pathway |
| SEKI INFLAMMATORY RESPONSE LPS DN | 23 | 16 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR UP | 293 | 203 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 11 | 36 | 24 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-29 | 803 | 810 | 1A,m8 | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| miR-326 | 1481 | 1488 | 1A,m8 | hsa-miR-326 | CCUCUGGGCCCUUCCUCCAG |
| hsa-miR-326 | CCUCUGGGCCCUUCCUCCAG | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.