Gene Page: PCBP1
Summary ?
| GeneID | 5093 |
| Symbol | PCBP1 |
| Synonyms | HEL-S-85|HNRPE1|HNRPX|hnRNP-E1|hnRNP-X |
| Description | poly(rC) binding protein 1 |
| Reference | MIM:601209|HGNC:HGNC:8647|Ensembl:ENSG00000169564|HPRD:03128|Vega:OTTHUMG00000129645 |
| Gene type | protein-coding |
| Map location | 2p13-p12 |
| Pascal p-value | 0.091 |
| Sherlock p-value | 0.011 |
| Fetal beta | 0.087 |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_TAP-PSD-95-CORE |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
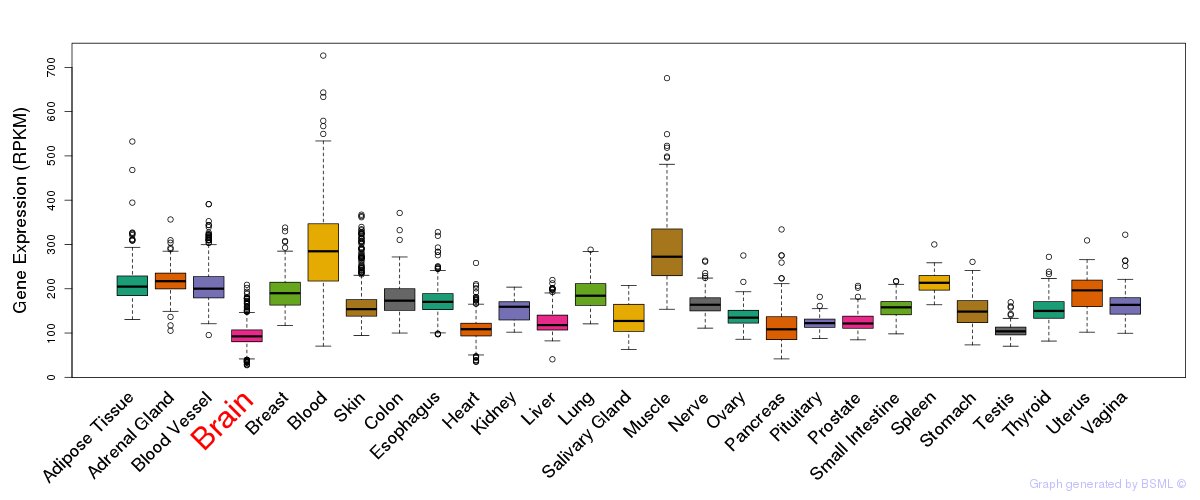
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NFIB | 0.90 | 0.80 |
| MYO5B | 0.89 | 0.79 |
| SOX5 | 0.89 | 0.84 |
| FEZF2 | 0.88 | 0.81 |
| ITSN1 | 0.87 | 0.76 |
| USP6NL | 0.87 | 0.80 |
| PELI2 | 0.87 | 0.72 |
| ZFPM2 | 0.86 | 0.76 |
| XPR1 | 0.86 | 0.77 |
| BCL11A | 0.86 | 0.72 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SERPINB6 | -0.53 | -0.60 |
| C5orf53 | -0.52 | -0.65 |
| FXYD1 | -0.50 | -0.71 |
| HLA-F | -0.50 | -0.61 |
| AF347015.27 | -0.49 | -0.70 |
| TNFSF12 | -0.49 | -0.48 |
| S100B | -0.49 | -0.68 |
| AF347015.31 | -0.49 | -0.71 |
| AIFM3 | -0.49 | -0.62 |
| HDDC2 | -0.49 | -0.48 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG SPLICEOSOME | 128 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | 140 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA PROCESSING | 161 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA SPLICING | 111 | 58 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| BIDUS METASTASIS UP | 214 | 134 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| RAHMAN TP53 TARGETS PHOSPHORYLATED | 21 | 18 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION DN | 337 | 230 | All SZGR 2.0 genes in this pathway |
| LEI MYB TARGETS | 318 | 215 | All SZGR 2.0 genes in this pathway |
| MARIADASON RESPONSE TO CURCUMIN SULINDAC 7 | 19 | 10 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 AND SATB1 UP | 87 | 52 | All SZGR 2.0 genes in this pathway |
| BOUDOUKHA BOUND BY IGF2BP2 | 111 | 59 | All SZGR 2.0 genes in this pathway |
| HOLLEMAN PREDNISOLONE RESISTANCE B ALL UP | 22 | 16 | All SZGR 2.0 genes in this pathway |
| HOLLEMAN PREDNISOLONE RESISTANCE ALL UP | 19 | 15 | All SZGR 2.0 genes in this pathway |