Gene Page: PDE1A
Summary ?
| GeneID | 5136 |
| Symbol | PDE1A |
| Synonyms | CAM-PDE-1A|HCAM-1|HCAM1|HSPDE1A |
| Description | phosphodiesterase 1A |
| Reference | MIM:171890|HGNC:HGNC:8774|Ensembl:ENSG00000115252|HPRD:01387|Vega:OTTHUMG00000132596 |
| Gene type | protein-coding |
| Map location | 2q32.1 |
| Pascal p-value | 0.025 |
| Sherlock p-value | 0.414 |
| Fetal beta | 0.102 |
| eGene | Caudate basal ganglia Cerebellar Hemisphere Hippocampus Nucleus accumbens basal ganglia Putamen basal ganglia Myers' cis & trans |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs940335 | chr7 | 99159332 | PDE1A | 5136 | 0.17 | trans | ||
| rs10931021 | 2 | 183368517 | PDE1A | ENSG00000115252.14 | 7.612E-7 | 0 | 19402 | gtex_brain_putamen_basal |
| rs2368301 | 2 | 183369652 | PDE1A | ENSG00000115252.14 | 6.274E-8 | 0 | 18267 | gtex_brain_putamen_basal |
| rs9789764 | 2 | 183373914 | PDE1A | ENSG00000115252.14 | 3.683E-6 | 0 | 14005 | gtex_brain_putamen_basal |
| rs6433976 | 2 | 183376081 | PDE1A | ENSG00000115252.14 | 4.07E-7 | 0 | 11838 | gtex_brain_putamen_basal |
| rs7595111 | 2 | 183376369 | PDE1A | ENSG00000115252.14 | 4.07E-7 | 0 | 11550 | gtex_brain_putamen_basal |
| rs10175845 | 2 | 183378125 | PDE1A | ENSG00000115252.14 | 3.501E-6 | 0 | 9794 | gtex_brain_putamen_basal |
| rs2082135 | 2 | 183378257 | PDE1A | ENSG00000115252.14 | 4.047E-7 | 0 | 9662 | gtex_brain_putamen_basal |
| rs4666850 | 2 | 183378710 | PDE1A | ENSG00000115252.14 | 4.044E-7 | 0 | 9209 | gtex_brain_putamen_basal |
| rs1991596 | 2 | 183379073 | PDE1A | ENSG00000115252.14 | 3.21E-6 | 0 | 8846 | gtex_brain_putamen_basal |
| rs2887207 | 2 | 183380721 | PDE1A | ENSG00000115252.14 | 5.189E-7 | 0 | 7198 | gtex_brain_putamen_basal |
| rs934261 | 2 | 183382345 | PDE1A | ENSG00000115252.14 | 3.897E-7 | 0 | 5574 | gtex_brain_putamen_basal |
| rs10174383 | 2 | 183382731 | PDE1A | ENSG00000115252.14 | 3.706E-6 | 0 | 5188 | gtex_brain_putamen_basal |
| rs1430149 | 2 | 183383268 | PDE1A | ENSG00000115252.14 | 3.86E-7 | 0 | 4651 | gtex_brain_putamen_basal |
| rs720245 | 2 | 183390144 | PDE1A | ENSG00000115252.14 | 3.732E-7 | 0 | -2225 | gtex_brain_putamen_basal |
| rs1835340 | 2 | 183393811 | PDE1A | ENSG00000115252.14 | 3.781E-7 | 0 | -5892 | gtex_brain_putamen_basal |
| rs4580353 | 2 | 183395232 | PDE1A | ENSG00000115252.14 | 1.46E-8 | 0 | -7313 | gtex_brain_putamen_basal |
| rs1430151 | 2 | 183395511 | PDE1A | ENSG00000115252.14 | 3.902E-9 | 0 | -7592 | gtex_brain_putamen_basal |
| rs68098724 | 2 | 183395631 | PDE1A | ENSG00000115252.14 | 3.902E-9 | 0 | -7712 | gtex_brain_putamen_basal |
| rs13027181 | 2 | 183395815 | PDE1A | ENSG00000115252.14 | 3.9E-9 | 0 | -7896 | gtex_brain_putamen_basal |
| rs13026884 | 2 | 183395835 | PDE1A | ENSG00000115252.14 | 3.9E-9 | 0 | -7916 | gtex_brain_putamen_basal |
| rs13026896 | 2 | 183395871 | PDE1A | ENSG00000115252.14 | 3.9E-9 | 0 | -7952 | gtex_brain_putamen_basal |
| rs397816195 | 2 | 183396838 | PDE1A | ENSG00000115252.14 | 6.306E-9 | 0 | -8919 | gtex_brain_putamen_basal |
| rs12693309 | 2 | 183397510 | PDE1A | ENSG00000115252.14 | 3.808E-9 | 0 | -9591 | gtex_brain_putamen_basal |
| rs12693310 | 2 | 183397531 | PDE1A | ENSG00000115252.14 | 2.054E-8 | 0 | -9612 | gtex_brain_putamen_basal |
| rs12693311 | 2 | 183397542 | PDE1A | ENSG00000115252.14 | 1.152E-8 | 0 | -9623 | gtex_brain_putamen_basal |
| rs11686003 | 2 | 183397980 | PDE1A | ENSG00000115252.14 | 3.266E-9 | 0 | -10061 | gtex_brain_putamen_basal |
| rs1430152 | 2 | 183398404 | PDE1A | ENSG00000115252.14 | 3.898E-9 | 0 | -10485 | gtex_brain_putamen_basal |
| rs35418011 | 2 | 183398774 | PDE1A | ENSG00000115252.14 | 3.898E-9 | 0 | -10855 | gtex_brain_putamen_basal |
| rs12162422 | 2 | 183398896 | PDE1A | ENSG00000115252.14 | 3.898E-9 | 0 | -10977 | gtex_brain_putamen_basal |
| rs11449946 | 2 | 183399798 | PDE1A | ENSG00000115252.14 | 6.769E-7 | 0 | -11879 | gtex_brain_putamen_basal |
| rs35385135 | 2 | 183400076 | PDE1A | ENSG00000115252.14 | 1.35E-8 | 0 | -12157 | gtex_brain_putamen_basal |
| rs11689324 | 2 | 183400185 | PDE1A | ENSG00000115252.14 | 1.458E-8 | 0 | -12266 | gtex_brain_putamen_basal |
| rs12473783 | 2 | 183400661 | PDE1A | ENSG00000115252.14 | 3.898E-9 | 0 | -12742 | gtex_brain_putamen_basal |
| rs10182788 | 2 | 183400730 | PDE1A | ENSG00000115252.14 | 1.53E-13 | 0 | -12811 | gtex_brain_putamen_basal |
| rs1583113 | 2 | 183401353 | PDE1A | ENSG00000115252.14 | 3.898E-9 | 0 | -13434 | gtex_brain_putamen_basal |
| rs1430153 | 2 | 183401673 | PDE1A | ENSG00000115252.14 | 1.53E-13 | 0 | -13754 | gtex_brain_putamen_basal |
| rs1430154 | 2 | 183401789 | PDE1A | ENSG00000115252.14 | 1.53E-13 | 0 | -13870 | gtex_brain_putamen_basal |
| rs7570760 | 2 | 183402841 | PDE1A | ENSG00000115252.14 | 3.898E-9 | 0 | -14922 | gtex_brain_putamen_basal |
| rs1430155 | 2 | 183403506 | PDE1A | ENSG00000115252.14 | 3.546E-7 | 0 | -15587 | gtex_brain_putamen_basal |
| rs10931023 | 2 | 183403962 | PDE1A | ENSG00000115252.14 | 4.51E-10 | 0 | -16043 | gtex_brain_putamen_basal |
| rs1965089 | 2 | 183404968 | PDE1A | ENSG00000115252.14 | 7.045E-11 | 0 | -17049 | gtex_brain_putamen_basal |
| rs7598587 | 2 | 183407212 | PDE1A | ENSG00000115252.14 | 1.458E-8 | 0 | -19293 | gtex_brain_putamen_basal |
| rs2368281 | 2 | 183408260 | PDE1A | ENSG00000115252.14 | 3.083E-7 | 0 | -20341 | gtex_brain_putamen_basal |
| rs934262 | 2 | 183409319 | PDE1A | ENSG00000115252.14 | 7.08E-8 | 0 | -21400 | gtex_brain_putamen_basal |
| rs934263 | 2 | 183409320 | PDE1A | ENSG00000115252.14 | 7.113E-8 | 0 | -21401 | gtex_brain_putamen_basal |
| rs6747954 | 2 | 183415490 | PDE1A | ENSG00000115252.14 | 4.514E-12 | 0 | -27571 | gtex_brain_putamen_basal |
| rs6711313 | 2 | 183416697 | PDE1A | ENSG00000115252.14 | 1.53E-13 | 0 | -28778 | gtex_brain_putamen_basal |
| rs4666588 | 2 | 183417707 | PDE1A | ENSG00000115252.14 | 4.417E-13 | 0 | -29788 | gtex_brain_putamen_basal |
| rs1430156 | 2 | 183423637 | PDE1A | ENSG00000115252.14 | 1.458E-8 | 0 | -35718 | gtex_brain_putamen_basal |
| rs4666589 | 2 | 183424775 | PDE1A | ENSG00000115252.14 | 2.514E-7 | 0 | -36856 | gtex_brain_putamen_basal |
| rs111326379 | 2 | 183425701 | PDE1A | ENSG00000115252.14 | 1.144E-11 | 0 | -37782 | gtex_brain_putamen_basal |
| rs139837068 | 2 | 183428054 | PDE1A | ENSG00000115252.14 | 1.282E-8 | 0 | -40135 | gtex_brain_putamen_basal |
| rs7558348 | 2 | 183428186 | PDE1A | ENSG00000115252.14 | 2.633E-6 | 0 | -40267 | gtex_brain_putamen_basal |
| rs10931024 | 2 | 183441026 | PDE1A | ENSG00000115252.14 | 1.503E-6 | 0 | -53107 | gtex_brain_putamen_basal |
| rs12479286 | 2 | 183441218 | PDE1A | ENSG00000115252.14 | 7.34E-7 | 0 | -53299 | gtex_brain_putamen_basal |
| rs7604873 | 2 | 183441726 | PDE1A | ENSG00000115252.14 | 6.309E-11 | 0 | -53807 | gtex_brain_putamen_basal |
| rs6758080 | 2 | 183442443 | PDE1A | ENSG00000115252.14 | 7.34E-7 | 0 | -54524 | gtex_brain_putamen_basal |
| rs7570144 | 2 | 183443136 | PDE1A | ENSG00000115252.14 | 7.34E-7 | 0 | -55217 | gtex_brain_putamen_basal |
| rs1527878 | 2 | 183446535 | PDE1A | ENSG00000115252.14 | 1.394E-9 | 0 | -58616 | gtex_brain_putamen_basal |
| rs1918700 | 2 | 183449725 | PDE1A | ENSG00000115252.14 | 8.161E-11 | 0 | -61806 | gtex_brain_putamen_basal |
| rs4490143 | 2 | 183450403 | PDE1A | ENSG00000115252.14 | 4.857E-10 | 0 | -62484 | gtex_brain_putamen_basal |
| rs2368330 | 2 | 183453542 | PDE1A | ENSG00000115252.14 | 2.561E-6 | 0 | -65623 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
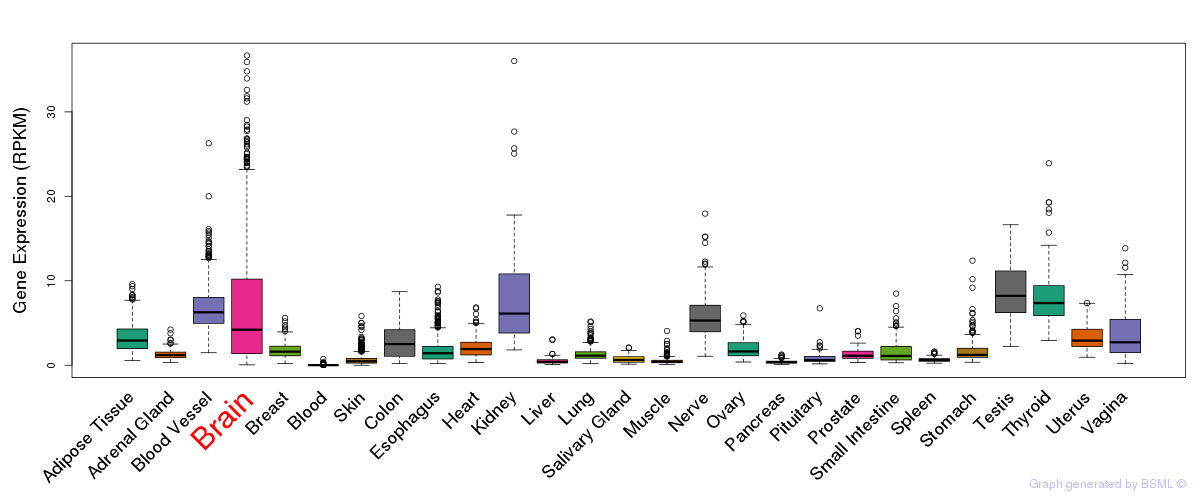
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| UGT8 | 0.96 | 0.94 |
| PLP1 | 0.95 | 0.96 |
| HSPA2 | 0.94 | 0.94 |
| GLDN | 0.94 | 0.91 |
| TF | 0.94 | 0.95 |
| CNDP1 | 0.94 | 0.89 |
| SLC31A2 | 0.94 | 0.78 |
| ABCA8 | 0.93 | 0.89 |
| KLK6 | 0.93 | 0.94 |
| FOLH1 | 0.93 | 0.88 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NR2C2AP | -0.53 | -0.78 |
| NKIRAS2 | -0.53 | -0.68 |
| TRNAU1AP | -0.52 | -0.73 |
| TBC1D10A | -0.52 | -0.58 |
| TUBB2B | -0.52 | -0.80 |
| ALKBH2 | -0.52 | -0.76 |
| PLEKHO1 | -0.52 | -0.77 |
| KIAA1949 | -0.51 | -0.74 |
| CCDC28B | -0.51 | -0.83 |
| AC009133.1 | -0.51 | -0.69 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PURINE METABOLISM | 159 | 96 | All SZGR 2.0 genes in this pathway |
| KEGG CALCIUM SIGNALING PATHWAY | 178 | 134 | All SZGR 2.0 genes in this pathway |
| KEGG TASTE TRANSDUCTION | 52 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME DAG AND IP3 SIGNALING | 32 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB2 | 101 | 78 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY EGFR IN CANCER | 109 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | 137 | 105 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR IN DISEASE | 127 | 88 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME OPIOID SIGNALLING | 78 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME CA DEPENDENT EVENTS | 30 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME PLC BETA MEDIATED EVENTS | 43 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY PDGF | 122 | 93 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | 95 | 76 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA S SIGNALLING EVENTS | 121 | 82 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNALING OF ACTIVATED FGFR | 100 | 74 | All SZGR 2.0 genes in this pathway |
| REACTOME PHOSPHOLIPASE C MEDIATED CASCADE | 54 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME CGMP EFFECTS | 19 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | 25 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET HOMEOSTASIS | 78 | 49 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR | 112 | 80 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL UP | 316 | 190 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 DN | 378 | 231 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B | 392 | 251 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL SHORT TERM | 32 | 15 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS DN | 314 | 188 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR DN | 191 | 123 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K27ME3 UP | 295 | 149 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| WU ALZHEIMER DISEASE DN | 19 | 12 | All SZGR 2.0 genes in this pathway |
| COLINA TARGETS OF 4EBP1 AND 4EBP2 | 356 | 214 | All SZGR 2.0 genes in this pathway |
| CROONQUIST IL6 DEPRIVATION UP | 20 | 10 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC ICP WITH H3K4ME3 | 445 | 257 | All SZGR 2.0 genes in this pathway |
| NIELSEN GIST | 98 | 66 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR DN | 244 | 157 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS UP | 279 | 155 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 | 227 | 149 | All SZGR 2.0 genes in this pathway |
| DURAND STROMA S UP | 297 | 194 | All SZGR 2.0 genes in this pathway |