Gene Page: PDGFRB
Summary ?
| GeneID | 5159 |
| Symbol | PDGFRB |
| Synonyms | CD140B|IBGC4|IMF1|JTK12|KOGS|PDGFR|PDGFR-1|PDGFR1|PENTT |
| Description | platelet derived growth factor receptor beta |
| Reference | MIM:173410|HGNC:HGNC:8804|Ensembl:ENSG00000113721|HPRD:01423|Vega:OTTHUMG00000130053 |
| Gene type | protein-coding |
| Map location | 5q33.1 |
| Pascal p-value | 0.384 |
| Sherlock p-value | 0.138 |
| Fetal beta | -0.07 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0032 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00459 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01718 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4501739 | chrX | 35960848 | PDGFRB | 5159 | 0.16 | trans |
Section II. Transcriptome annotation
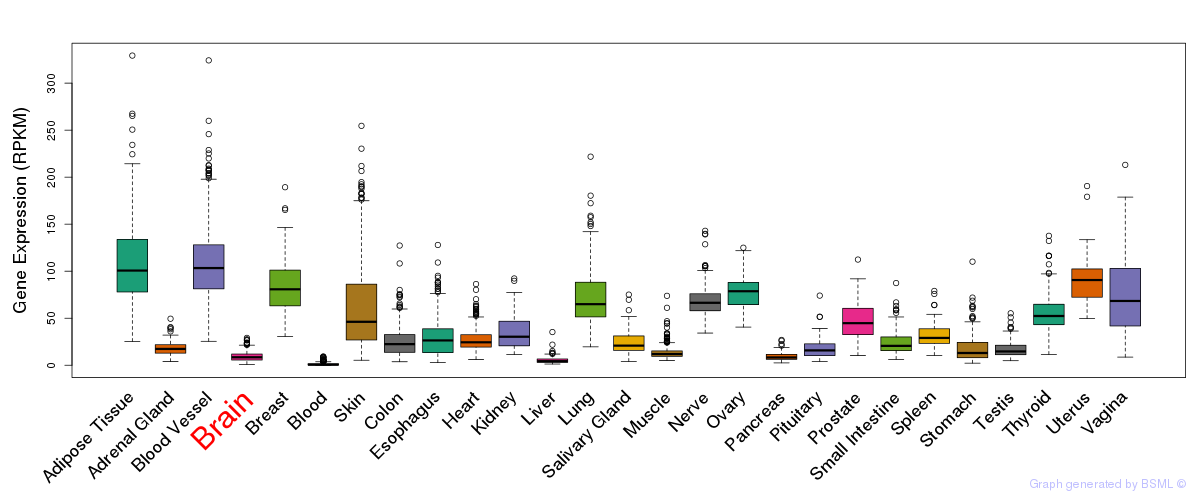
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PRDX3 | 0.79 | 0.78 |
| POP4 | 0.78 | 0.75 |
| TMEM111 | 0.78 | 0.76 |
| GLO1 | 0.78 | 0.78 |
| MOCS2 | 0.77 | 0.79 |
| PEX19 | 0.76 | 0.75 |
| SNX17 | 0.76 | 0.77 |
| OCIAD1 | 0.76 | 0.74 |
| CISD1 | 0.75 | 0.75 |
| TMX2 | 0.75 | 0.73 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AC010300.1 | -0.44 | -0.45 |
| AF347015.18 | -0.43 | -0.23 |
| HSP90AB4P | -0.43 | -0.44 |
| C10orf108 | -0.41 | -0.37 |
| AC005921.3 | -0.40 | -0.48 |
| AC100783.1 | -0.40 | -0.33 |
| FAM159B | -0.37 | -0.33 |
| AL031587.2 | -0.37 | -0.30 |
| AC135724.1 | -0.37 | -0.40 |
| ZNF814 | -0.35 | -0.39 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005021 | vascular endothelial growth factor receptor activity | IEA | - | |
| GO:0005019 | platelet-derived growth factor beta-receptor activity | IDA | 2536956 | |
| GO:0004872 | receptor activity | IEA | - | |
| GO:0005161 | platelet-derived growth factor receptor binding | IPI | 2542288 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004992 | platelet activating factor receptor activity | TAS | 2536956 | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0048407 | platelet-derived growth factor binding | IPI | 2536956 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007165 | signal transduction | TAS | 2536956 |10821867 | |
| GO:0008284 | positive regulation of cell proliferation | IDA | 17470632 | |
| GO:0048008 | platelet-derived growth factor receptor signaling pathway | IDA | 2536956 | |
| GO:0030335 | positive regulation of cell migration | IDA | 17470632 | |
| GO:0050730 | regulation of peptidyl-tyrosine phosphorylation | IEA | - | |
| GO:0046777 | protein amino acid autophosphorylation | IDA | 2536956 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BAG1 | RAP46 | BCL2-associated athanogene | - | HPRD | 8947043 |
| CAV1 | CAV | MSTP085 | VIP21 | caveolin 1, caveolae protein, 22kDa | Affinity Capture-Western Co-fractionation Reconstituted Complex | BioGRID | 10066366 |
| CAV3 | LGMD1C | LQT9 | MGC126100 | MGC126101 | MGC126129 | VIP-21 | VIP21 | caveolin 3 | Reconstituted Complex | BioGRID | 10066366 |
| CBL | C-CBL | CBL2 | RNF55 | Cas-Br-M (murine) ecotropic retroviral transforming sequence | c-Cbl interacts with and ubiquitinates PDGF-R-beta. This interaction was modeled on a demonstrated interaction between human c-Cbl and PDGF-R-beta from an unspecified species. | BIND | 10514377 |
| COPA | FLJ26320 | HEP-COP | coatomer protein complex, subunit alpha | - | HPRD,BioGRID | 9207175 |
| COPB1 | COPB | DKFZp761K102 | FLJ10341 | coatomer protein complex, subunit beta 1 | - | HPRD | 9207175 |
| CRK | CRKII | v-crk sarcoma virus CT10 oncogene homolog (avian) | PDGFR interacts with Crk. This interaction was modelled on a demonstrated interaction between rat PDGFR and Crk from an unknown species. | BIND | 2173144 |
| CRK | CRKII | v-crk sarcoma virus CT10 oncogene homolog (avian) | - | HPRD,BioGRID | 10733900 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | - | HPRD,BioGRID | 9506992 |
| EIF2AK2 | EIF2AK1 | MGC126524 | PKR | PRKR | eukaryotic translation initiation factor 2-alpha kinase 2 | Affinity Capture-Western | BioGRID | 11350938 |
| EPHB1 | ELK | EPHT2 | FLJ37986 | Hek6 | NET | EPH receptor B1 | Elk phosphorylates PDGFR. This interaction was modelled on a demonstrated interaction between Elk from an unknown species and human PDGFR. | BIND | 7689724 |
| FYN | MGC45350 | SLK | SYN | FYN oncogene related to SRC, FGR, YES | - | HPRD | 1696179 |
| GRB10 | GRB-IR | Grb-10 | IRBP | KIAA0207 | MEG1 | RSS | growth factor receptor-bound protein 10 | - | HPRD | 9006901 |
| GRB14 | - | growth factor receptor-bound protein 14 | - | HPRD,BioGRID | 8647858 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | - | HPRD,BioGRID | 7935391 |
| ITGB3 | CD61 | GP3A | GPIIIa | integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) | Affinity Capture-Western | BioGRID | 10964931 |
| KRTAP4-12 | KAP4.12 | KRTAP4.12 | keratin associated protein 4-12 | Two-hybrid | BioGRID | 16189514 |
| NCK1 | MGC12668 | NCK | NCKalpha | NCK adaptor protein 1 | Autophosphorylated PDGFR interacts with Nck. This interaction was modelled on a demonstrated interaction between human PDGFR and Nck from an unspecified species. | BIND | 8890167 |
| NCK1 | MGC12668 | NCK | NCKalpha | NCK adaptor protein 1 | - | HPRD | 9362449 |
| NCK1 | MGC12668 | NCK | NCKalpha | NCK adaptor protein 1 | Affinity Capture-Western | BioGRID | 1333047 |10026169 |
| NCK2 | GRB4 | NCKbeta | NCK adaptor protein 2 | - | HPRD,BioGRID | 11027258 |
| PDAP1 | HASPP28 | PAP | PAP1 | PDGFA associated protein 1 | - | HPRD | 11331882 |
| PDGFB | FLJ12858 | PDGF2 | SIS | SSV | c-sis | platelet-derived growth factor beta polypeptide (simian sarcoma viral (v-sis) oncogene homolog) | - | HPRD | 11903042 |
| PDGFD | IEGF | MGC26867 | MSTP036 | SCDGF-B | platelet derived growth factor D | - | HPRD | 14514732 |
| PDGFRA | CD140A | MGC74795 | PDGFR2 | Rhe-PDGFRA | platelet-derived growth factor receptor, alpha polypeptide | Affinity Capture-Western Reconstituted Complex | BioGRID | 2542288 |7523122 |
| PDGFRB | CD140B | JTK12 | PDGF-R-beta | PDGFR | PDGFR1 | platelet-derived growth factor receptor, beta polypeptide | Affinity Capture-Western Far Western | BioGRID | 7523122 |11046132 |
| PDGFRB | CD140B | JTK12 | PDGF-R-beta | PDGFR | PDGFR1 | platelet-derived growth factor receptor, beta polypeptide | PDGFRB homodimerizes and cross-phosphorylates. | BIND | 15889147 |
| PIK3R1 | GRB1 | p85 | p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | in vivo | BioGRID | 10697503 |
| PIK3R1 | GRB1 | p85 | p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | Beta-PDGFR interacts with p85. This interaction was modeled on a demonstrated interaction between human beta-PDGFR and rat or bovine p85. | BIND | 1372092 |7689724 |7876130 |
| PIK3R1 | GRB1 | p85 | p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | Phosphorylated beta-PDGFR interacts with p85-alpha. This interaction was modeled on a demonstrated interaction between human beta-PDGFR and bovine p85-alpha. | BIND | 8388538 |
| PIK3R2 | P85B | p85 | p85-BETA | phosphoinositide-3-kinase, regulatory subunit 2 (beta) | PDGFR interacts with p85-alpha. This interaction was modeled based on a demonstrated interaction between human PDGFR and p85-alpha from an unknown species. | BIND | 11172806 |
| PIK3R3 | DKFZp686P05226 | FLJ41892 | p55 | p55-GAMMA | phosphoinositide-3-kinase, regulatory subunit 3 (gamma) | PDGFR interacts with p55-gamma. This interaction was modeled based on a demonstrated interaction between human PDGFR and p55-gamma from an unknown species. | BIND | 11172806 |
| PLAUR | CD87 | UPAR | URKR | plasminogen activator, urokinase receptor | An unspecified isoform of PLAUR (uPAR) interacts with PDGFRB. | BIND | 15889147 |
| PLCG1 | PLC-II | PLC1 | PLC148 | PLCgamma1 | phospholipase C, gamma 1 | - | HPRD,BioGRID | 8443409 |
| PLCG1 | PLC-II | PLC1 | PLC148 | PLCgamma1 | phospholipase C, gamma 1 | Autophosphorylated PDGFR interacts with PLC-gamma. This interaction was modelled on a demonstrated interaction between human PDGFR and PLC-gamma from an unspecified species. | BIND | 8890167 |
| PLCG1 | PLC-II | PLC1 | PLC148 | PLCgamma1 | phospholipase C, gamma 1 | Phosphorylated beta-PDGFR interacts with PLC-gamma. This interaction was modeled on a demonstrated interaction between human beta-PDGFR and bovine PLC-gamma. | BIND | 8388538 |
| PLCG1 | PLC-II | PLC1 | PLC148 | PLCgamma1 | phospholipase C, gamma 1 | Beta-PDGFR interacts with PLC-gamma 1. This interaction was modeled on a demonstrated interaction between human beta-PDGFR and PLC-gamma from bovine and unspecified sources. | BIND | 2173144 |7592796 |7689724 |7876130 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | - | HPRD | 10806474 |
| PTPN11 | BPTP3 | CFC | MGC14433 | NS1 | PTP-1D | PTP2C | SH-PTP2 | SH-PTP3 | SHP2 | protein tyrosine phosphatase, non-receptor type 11 | Reconstituted Complex Two-hybrid | BioGRID | 7691811 |11266449 |
| PTPN11 | BPTP3 | CFC | MGC14433 | NS1 | PTP-1D | PTP2C | SH-PTP2 | SH-PTP3 | SHP2 | protein tyrosine phosphatase, non-receptor type 11 | PDGFR interacts with Syp. | BIND | 7688466 |
| PTPN11 | BPTP3 | CFC | MGC14433 | NS1 | PTP-1D | PTP2C | SH-PTP2 | SH-PTP3 | SHP2 | protein tyrosine phosphatase, non-receptor type 11 | Autophosphorylated PDGFR interacts with Syp. This interaction was modelled on a demonstrated interaction between human PDGFR and Syp from an unspecified species. | BIND | 8890167 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | - | HPRD,BioGRID | 2475255 |
| RASA1 | CM-AVM | CMAVM | DKFZp434N071 | GAP | PKWS | RASA | RASGAP | p120GAP | p120RASGAP | RAS p21 protein activator (GTPase activating protein) 1 | Activated PDGFR interacts with GAP. This interaction was modeled on a demonstrated interaction between human PDGFR and canine GAP. | BIND | 2157284 |
| RASA1 | CM-AVM | CMAVM | DKFZp434N071 | GAP | PKWS | RASA | RASGAP | p120GAP | p120RASGAP | RAS p21 protein activator (GTPase activating protein) 1 | N-terminal SH2 domain of GAP interacts with PDGFR. This interaction was modeled on a demonstrated interaction between GAP from human and PDGFR from an unspecified species. | BIND | 1385407 |2173144 |7689724 |8344248 |
| RASA1 | CM-AVM | CMAVM | DKFZp434N071 | GAP | PKWS | RASA | RASGAP | p120GAP | p120RASGAP | RAS p21 protein activator (GTPase activating protein) 1 | - | HPRD,BioGRID | 10697503 |11896619 |
| RASA1 | CM-AVM | CMAVM | DKFZp434N071 | GAP | PKWS | RASA | RASGAP | p120GAP | p120RASGAP | RAS p21 protein activator (GTPase activating protein) 1 | Autophosphorylated PDGFR interacts with GAP. This interaction was modelled on a demonstrated interaction between human PDGFR and GAP from an unspecified species. | BIND | 8890167 |
| S1PR1 | CHEDG1 | D1S3362 | ECGF1 | EDG-1 | EDG1 | FLJ58121 | S1P1 | sphingosine-1-phosphate receptor 1 | - | HPRD,BioGRID | 12480944 |
| SH3KBP1 | CIN85 | GIG10 | MIG18 | SH3-domain kinase binding protein 1 | - | HPRD,BioGRID | 12177062 |
| SHB | RP11-3J10.8 | bA3J10.2 | Src homology 2 domain containing adaptor protein B | Shb interacts with the PDGF beta-receptor. | BIND | 7537362 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | PDGFR interacts with p52Shc | BIND | 8195171 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | PDGFR interacts with p66Shc. | BIND | 8195171 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | - | HPRD,BioGRID | 8195171 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | Autophosphorylated PDGFR interacts with SHC. This interaction was modelled on a demonstrated interaction between human PDGFR and SHC from an unspecified species. | BIND | 8890167 |
| SLC9A3R1 | EBP50 | NHERF | NHERF1 | NPHLOP2 | solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1 | - | HPRD,BioGRID | 11046132 |
| SNX1 | HsT17379 | MGC8664 | SNX1A | Vps5 | sorting nexin 1 | - | HPRD,BioGRID | 9819414 |
| SNX2 | MGC5204 | TRG-9 | sorting nexin 2 | Affinity Capture-Western | BioGRID | 9819414 |
| SNX4 | - | sorting nexin 4 | Affinity Capture-Western | BioGRID | 9819414 |
| SOCS1 | CIS1 | CISH1 | JAB | SOCS-1 | SSI-1 | SSI1 | TIP3 | suppressor of cytokine signaling 1 | Socs1 interacts with PDGF-R. This interaction was modeled on a demonstrated interaction between mouse Socs1 and human PDGF-R. | BIND | 10022833 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | PDGFR interacts with Src. This interaction was modelled on a demonstrated interaction between rat PDGFR and Src from an unknown species. | BIND | 2173144 |
| VAV1 | VAV | vav 1 guanine nucleotide exchange factor | Affinity Capture-Western | BioGRID | 10938113 |
| VAV2 | - | vav 2 guanine nucleotide exchange factor | Affinity Capture-Western | BioGRID | 10938113 |
| VAV3 | FLJ40431 | vav 3 guanine nucleotide exchange factor | Affinity Capture-Western | BioGRID | 10938113 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG CALCIUM SIGNALING PATHWAY | 178 | 134 | All SZGR 2.0 genes in this pathway |
| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION | 267 | 161 | All SZGR 2.0 genes in this pathway |
| KEGG FOCAL ADHESION | 201 | 138 | All SZGR 2.0 genes in this pathway |
| KEGG GAP JUNCTION | 90 | 68 | All SZGR 2.0 genes in this pathway |
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG GLIOMA | 65 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG PROSTATE CANCER | 89 | 75 | All SZGR 2.0 genes in this pathway |
| KEGG MELANOMA | 71 | 57 | All SZGR 2.0 genes in this pathway |
| PID PTP1B PATHWAY | 52 | 40 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN3 PATHWAY | 43 | 25 | All SZGR 2.0 genes in this pathway |
| PID S1P S1P3 PATHWAY | 29 | 24 | All SZGR 2.0 genes in this pathway |
| PID NECTIN PATHWAY | 30 | 20 | All SZGR 2.0 genes in this pathway |
| PID TCPTP PATHWAY | 43 | 33 | All SZGR 2.0 genes in this pathway |
| PID SHP2 PATHWAY | 58 | 46 | All SZGR 2.0 genes in this pathway |
| PID S1P S1P1 PATHWAY | 21 | 18 | All SZGR 2.0 genes in this pathway |
| PID UPA UPAR PATHWAY | 42 | 30 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PID MYC REPRESS PATHWAY | 63 | 52 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY PDGF | 122 | 93 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | 95 | 76 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE UP | 351 | 230 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 48HR UP | 128 | 95 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 96HR UP | 117 | 84 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 DN | 378 | 231 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND SERUM DEPRIVATION UP | 211 | 136 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| CAIRO PML TARGETS BOUND BY MYC DN | 14 | 12 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| HAEGERSTRAND RESPONSE TO IMATINIB | 9 | 5 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B | 392 | 251 | All SZGR 2.0 genes in this pathway |
| MYLLYKANGAS AMPLIFICATION HOT SPOT 27 | 15 | 8 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| TERAMOTO OPN TARGETS CLUSTER 6 | 27 | 17 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| JAZAG TGFB1 SIGNALING VIA SMAD4 DN | 66 | 38 | All SZGR 2.0 genes in this pathway |
| SASAI RESISTANCE TO NEOPLASTIC TRANSFROMATION | 50 | 31 | All SZGR 2.0 genes in this pathway |
| THEODOROU MAMMARY TUMORIGENESIS | 31 | 24 | All SZGR 2.0 genes in this pathway |
| WEI MIR34A TARGETS | 148 | 97 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| JECHLINGER EPITHELIAL TO MESENCHYMAL TRANSITION UP | 71 | 51 | All SZGR 2.0 genes in this pathway |
| ABRAHAM ALPC VS MULTIPLE MYELOMA UP | 26 | 22 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| JAZAERI BREAST CANCER BRCA1 VS BRCA2 DN | 43 | 31 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 24HR DN | 148 | 102 | All SZGR 2.0 genes in this pathway |
| CHIBA RESPONSE TO TSA UP | 52 | 33 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| GERHOLD ADIPOGENESIS DN | 64 | 44 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR DN | 178 | 121 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS PEAK AT 0HR | 63 | 48 | All SZGR 2.0 genes in this pathway |
| JI CARCINOGENESIS BY KRAS AND STK11 DN | 17 | 12 | All SZGR 2.0 genes in this pathway |
| CLAUS PGR POSITIVE MENINGIOMA DN | 12 | 10 | All SZGR 2.0 genes in this pathway |
| MISHRA CARCINOMA ASSOCIATED FIBROBLAST UP | 24 | 14 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K27ME3 DN | 228 | 114 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS DN | 138 | 92 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC | 420 | 269 | All SZGR 2.0 genes in this pathway |
| RUIZ TNC TARGETS UP | 153 | 107 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| YAMASHITA LIVER CANCER STEM CELL UP | 47 | 38 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF LCP WITH H3K4ME3 | 128 | 68 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS LCP WITH H3K4ME3 | 174 | 100 | All SZGR 2.0 genes in this pathway |
| NAKAYAMA FGF2 TARGETS | 29 | 17 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN | 253 | 192 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR BOUND ES | 462 | 273 | All SZGR 2.0 genes in this pathway |
| ANASTASSIOU CANCER MESENCHYMAL TRANSITION SIGNATURE | 64 | 40 | All SZGR 2.0 genes in this pathway |
| DURAND STROMA S UP | 297 | 194 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-29 | 1907 | 1913 | m8 | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| miR-30-5p | 1784 | 1790 | 1A | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-378 | 1123 | 1129 | 1A | hsa-miR-378 | CUCCUGACUCCAGGUCCUGUGU |
| miR-9 | 1763 | 1770 | 1A,m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.