Gene Page: CDK5RAP1
Summary ?
| GeneID | 51654 |
| Symbol | CDK5RAP1 |
| Synonyms | C20orf34|C42|CGI-05|HSPC167 |
| Description | CDK5 regulatory subunit associated protein 1 |
| Reference | MIM:608200|HGNC:HGNC:15880|Ensembl:ENSG00000101391|HPRD:12189|Vega:OTTHUMG00000032256 |
| Gene type | protein-coding |
| Map location | 20q11.21 |
| Pascal p-value | 0.208 |
| Sherlock p-value | 0.003 |
| Fetal beta | 0.321 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 3 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg26688574 | 20 | 31989989 | CDK5RAP1 | 1.291E-4 | 0.377 | 0.03 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs12527253 | chr6 | 89293744 | CDK5RAP1 | 51654 | 0.06 | trans | ||
| rs687928 | chr11 | 121996260 | CDK5RAP1 | 51654 | 0.16 | trans |
Section II. Transcriptome annotation
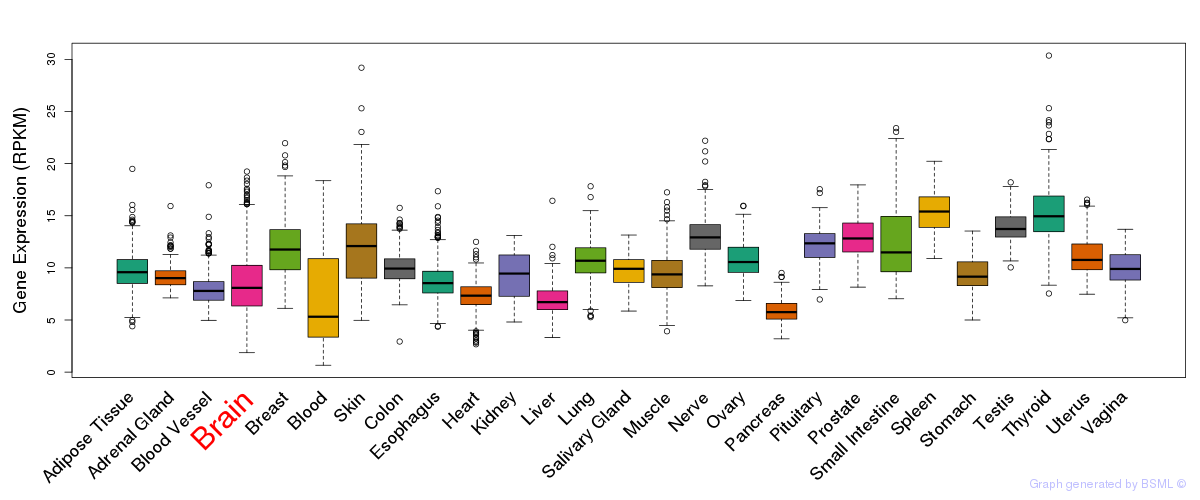
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0042808 | neuronal Cdc2-like kinase binding | NAS | neuron (GO term level: 7) | 10721722 |
| GO:0003824 | catalytic activity | IEA | - | |
| GO:0005506 | iron ion binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 15231747 | |
| GO:0046872 | metal ion binding | IEA | - | |
| GO:0051539 | 4 iron, 4 sulfur cluster binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007420 | brain development | NAS | Brain (GO term level: 7) | 10915792 |
| GO:0045664 | regulation of neuron differentiation | NAS | neuron, neurogenesis (GO term level: 9) | 10721722 |
| GO:0045736 | negative regulation of cyclin-dependent protein kinase activity | IDA | 11882646 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005575 | cellular_component | ND | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| GARY CD5 TARGETS DN | 431 | 263 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| BREDEMEYER RAG SIGNALING VIA ATM NOT VIA NFKB DN | 38 | 23 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |