Gene Page: PFKFB4
Summary ?
| GeneID | 5210 |
| Symbol | PFKFB4 |
| Synonyms | - |
| Description | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| Reference | MIM:605320|HGNC:HGNC:8875|Ensembl:ENSG00000114268|HPRD:05613|Vega:OTTHUMG00000133528 |
| Gene type | protein-coding |
| Map location | 3p22-p21 |
| Pascal p-value | 0.181 |
| Sherlock p-value | 0.812 |
| DEG p-value | DEG:Zhao_2015:p=2.09e-04:q=0.0860 |
| Fetal beta | 1.125 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DEG:Zhao_2015 | RNA Sequencing analysis | Transcriptome sequencing and genome-wide association analyses reveal lysosomal function and actin cytoskeleton remodeling in schizophrenia and bipolar disorder. |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
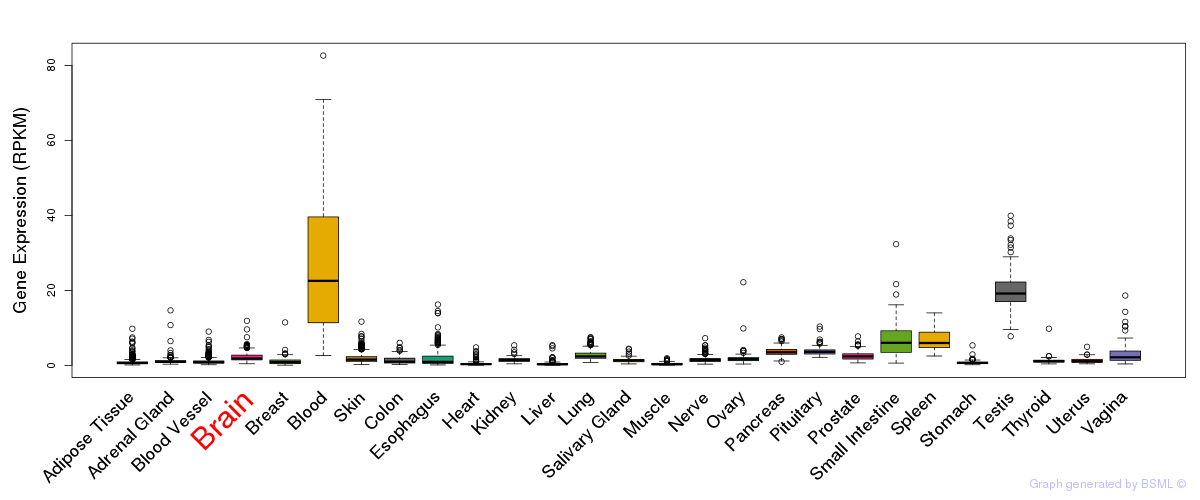
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG FRUCTOSE AND MANNOSE METABOLISM | 34 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME GLYCOLYSIS | 29 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF CARBOHYDRATES | 247 | 154 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUCOSE METABOLISM | 69 | 42 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA UP | 92 | 57 | All SZGR 2.0 genes in this pathway |
| HOOI ST7 TARGETS DN | 123 | 78 | All SZGR 2.0 genes in this pathway |
| OXFORD RALA TARGETS DN | 10 | 8 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| LIAO HAVE SOX4 BINDING SITES | 40 | 26 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS | 539 | 324 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO DN | 200 | 112 | All SZGR 2.0 genes in this pathway |
| MENSE HYPOXIA UP | 98 | 71 | All SZGR 2.0 genes in this pathway |
| LEONARD HYPOXIA | 47 | 35 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA NORMAL | 311 | 205 | All SZGR 2.0 genes in this pathway |
| XU GH1 EXOGENOUS TARGETS DN | 120 | 69 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS DN | 145 | 93 | All SZGR 2.0 genes in this pathway |
| LIN NPAS4 TARGETS UP | 163 | 100 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA METAGENE | 242 | 168 | All SZGR 2.0 genes in this pathway |
| MUELLER COMMON TARGETS OF AML FUSIONS DN | 31 | 21 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR DN | 88 | 59 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS UP | 217 | 131 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS GROUP1 | 136 | 76 | All SZGR 2.0 genes in this pathway |
| FARDIN HYPOXIA 11 | 32 | 29 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 AND SATB1 UP | 87 | 52 | All SZGR 2.0 genes in this pathway |
| YANG BCL3 TARGETS UP | 364 | 236 | All SZGR 2.0 genes in this pathway |
| LIU IL13 MEMORY MODEL UP | 17 | 12 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |
| KUMAR PATHOGEN LOAD BY MACROPHAGES | 275 | 155 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |