Gene Page: PHKG2
Summary ?
| GeneID | 5261 |
| Symbol | PHKG2 |
| Synonyms | GSD9C |
| Description | phosphorylase kinase, gamma 2 (testis) |
| Reference | MIM:172471|HGNC:HGNC:8931|Ensembl:ENSG00000156873|HPRD:01405|Vega:OTTHUMG00000132400 |
| Gene type | protein-coding |
| Map location | 16p11.2 |
| Pascal p-value | 0.034 |
| Sherlock p-value | 0.008 |
| Fetal beta | -0.633 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01775 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg02091607 | 16 | 30760815 | PHKG2 | 5.49E-4 | 0.282 | 0.049 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
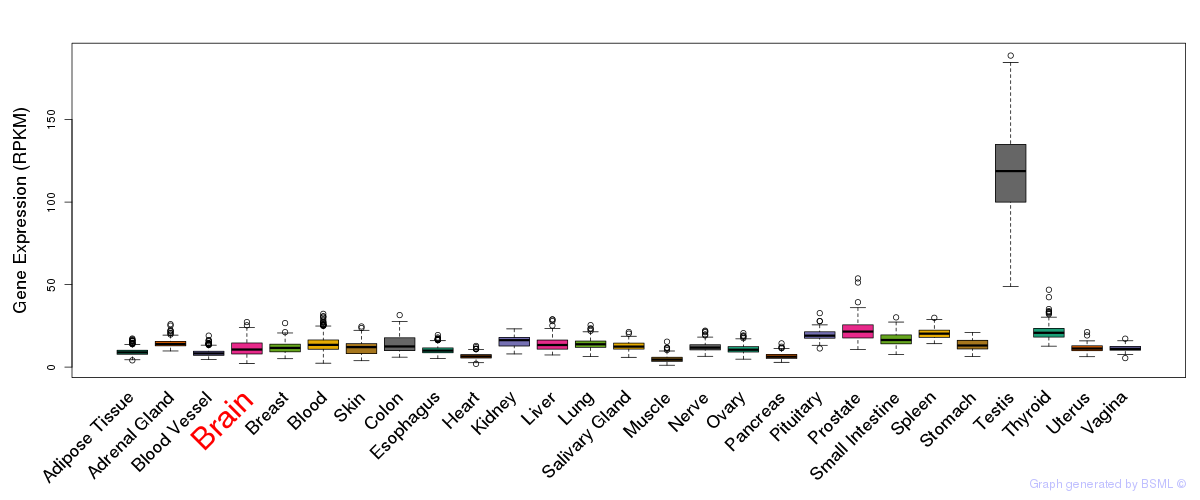
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CABLES2 | 0.91 | 0.94 |
| PTPRS | 0.90 | 0.93 |
| USP22 | 0.90 | 0.93 |
| C17orf63 | 0.89 | 0.91 |
| ARID1B | 0.89 | 0.91 |
| PHF12 | 0.89 | 0.90 |
| FOXK2 | 0.89 | 0.92 |
| ZNF142 | 0.89 | 0.91 |
| YTHDF1 | 0.89 | 0.92 |
| TGFBRAP1 | 0.88 | 0.93 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.71 | -0.82 |
| MT-CO2 | -0.69 | -0.83 |
| AF347015.27 | -0.68 | -0.80 |
| C5orf53 | -0.66 | -0.71 |
| MT-CYB | -0.66 | -0.78 |
| FXYD1 | -0.66 | -0.77 |
| AF347015.33 | -0.66 | -0.75 |
| IFI27 | -0.66 | -0.77 |
| AF347015.8 | -0.66 | -0.80 |
| HIGD1B | -0.66 | -0.80 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005516 | calmodulin binding | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0005524 | ATP binding | NAS | - | |
| GO:0004672 | protein kinase activity | IEA | - | |
| GO:0004674 | protein serine/threonine kinase activity | NAS | - | |
| GO:0004689 | phosphorylase kinase activity | NAS | 8020963 | |
| GO:0004689 | phosphorylase kinase activity | TAS | 2948189 | |
| GO:0016740 | transferase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0006468 | protein amino acid phosphorylation | NAS | - | |
| GO:0006091 | generation of precursor metabolites and energy | TAS | 8896567 | |
| GO:0005978 | glycogen biosynthetic process | IEA | - | |
| GO:0005975 | carbohydrate metabolic process | IEA | - | |
| GO:0008150 | biological_process | ND | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005575 | cellular_component | ND | - | |
| GO:0005964 | phosphorylase kinase complex | IEA | - | |
| GO:0005964 | phosphorylase kinase complex | NAS | 8020963 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CALCIUM SIGNALING PATHWAY | 178 | 134 | All SZGR 2.0 genes in this pathway |
| KEGG INSULIN SIGNALING PATHWAY | 137 | 103 | All SZGR 2.0 genes in this pathway |
| REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | 18 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF CARBOHYDRATES | 247 | 154 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUCOSE METABOLISM | 69 | 42 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN UP | 239 | 157 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| SU TESTIS | 76 | 53 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION NEOCORTEX DN | 88 | 58 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS DN | 141 | 92 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN | 270 | 181 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC ICP WITH H3K4ME3 | 445 | 257 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-1/206 | 57 | 63 | 1A | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.