Gene Page: PIK3C3
Summary ?
| GeneID | 5289 |
| Symbol | PIK3C3 |
| Synonyms | VPS34|Vps34|hVps34 |
| Description | phosphatidylinositol 3-kinase catalytic subunit type 3 |
| Reference | MIM:602609|HGNC:HGNC:8974|HPRD:04009| |
| Gene type | protein-coding |
| Map location | 18q12.3 |
| Pascal p-value | 0.123 |
| Sherlock p-value | 0.438 |
| Fetal beta | 0.429 |
| eGene | Cerebellum Cortex Nucleus accumbens basal ganglia Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 2 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
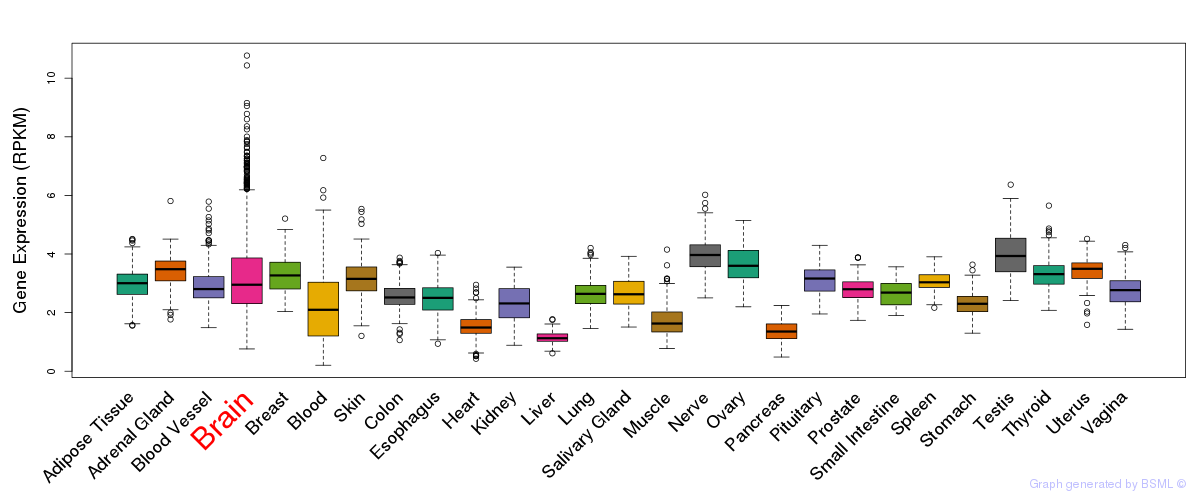
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CHN1 | 0.81 | 0.82 |
| RGS4 | 0.78 | 0.85 |
| TMEM155 | 0.76 | 0.69 |
| KCNK1 | 0.75 | 0.77 |
| ANXA7 | 0.75 | 0.78 |
| GCNT4 | 0.75 | 0.73 |
| ANXA11 | 0.75 | 0.82 |
| SLC39A8 | 0.75 | 0.82 |
| TPMT | 0.74 | 0.73 |
| THEMIS | 0.74 | 0.70 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SH3BP2 | -0.67 | -0.79 |
| KIAA1949 | -0.67 | -0.67 |
| PKN1 | -0.67 | -0.74 |
| TUBB2B | -0.66 | -0.74 |
| CCDC123 | -0.65 | -0.66 |
| ZNF311 | -0.65 | -0.66 |
| PDE9A | -0.65 | -0.75 |
| MARCKSL1 | -0.65 | -0.67 |
| GMIP | -0.64 | -0.61 |
| ZNF300 | -0.63 | -0.61 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 7504174 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004428 | inositol or phosphatidylinositol kinase activity | IEA | - | |
| GO:0016773 | phosphotransferase activity, alcohol group as acceptor | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0016303 | 1-phosphatidylinositol-3-kinase activity | TAS | 7628435 | |
| GO:0030145 | manganese ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0048015 | phosphoinositide-mediated signaling | IEA | - | |
| GO:0046854 | phosphoinositide phosphorylation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 14751759 | |
| GO:0005942 | phosphoinositide 3-kinase complex | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG INOSITOL PHOSPHATE METABOLISM | 54 | 42 | All SZGR 2.0 genes in this pathway |
| KEGG PHOSPHATIDYLINOSITOL SIGNALING SYSTEM | 76 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG REGULATION OF AUTOPHAGY | 35 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME INSULIN RECEPTOR SIGNALLING CASCADE | 87 | 64 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | 12 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | 17 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME PHOSPHOLIPID METABOLISM | 198 | 112 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | 10 | 7 | All SZGR 2.0 genes in this pathway |
| REACTOME PI METABOLISM | 48 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY INSULIN RECEPTOR | 108 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME TOLL RECEPTOR CASCADES | 118 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K CASCADE | 71 | 51 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| CHEMNITZ RESPONSE TO PROSTAGLANDIN E2 DN | 391 | 222 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL UP | 133 | 78 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS UP | 380 | 213 | All SZGR 2.0 genes in this pathway |
| GRANDVAUX IRF3 TARGETS DN | 19 | 14 | All SZGR 2.0 genes in this pathway |
| HEDVAT ELF4 TARGETS UP | 12 | 8 | All SZGR 2.0 genes in this pathway |
| ASTIER INTEGRIN SIGNALING | 59 | 44 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK DN | 318 | 220 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G6 | 153 | 112 | All SZGR 2.0 genes in this pathway |
| SARTIPY NORMAL AT INSULIN RESISTANCE DN | 21 | 11 | All SZGR 2.0 genes in this pathway |
| GU PDEF TARGETS UP | 71 | 49 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER DN | 238 | 145 | All SZGR 2.0 genes in this pathway |
| YAMASHITA LIVER CANCER STEM CELL UP | 47 | 38 | All SZGR 2.0 genes in this pathway |
| YAMASHITA LIVER CANCER STEM CELL DN | 76 | 51 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 12 | 79 | 54 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 UP | 344 | 215 | All SZGR 2.0 genes in this pathway |
| MIZUSHIMA AUTOPHAGOSOME FORMATION | 19 | 15 | All SZGR 2.0 genes in this pathway |