Gene Page: PLG
Summary ?
| GeneID | 5340 |
| Symbol | PLG |
| Synonyms | - |
| Description | plasminogen |
| Reference | MIM:173350|HGNC:HGNC:9071|Ensembl:ENSG00000122194|HPRD:01417|Vega:OTTHUMG00000015957 |
| Gene type | protein-coding |
| Map location | 6q26 |
| Pascal p-value | 0.037 |
| Fetal beta | 0.028 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0033 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
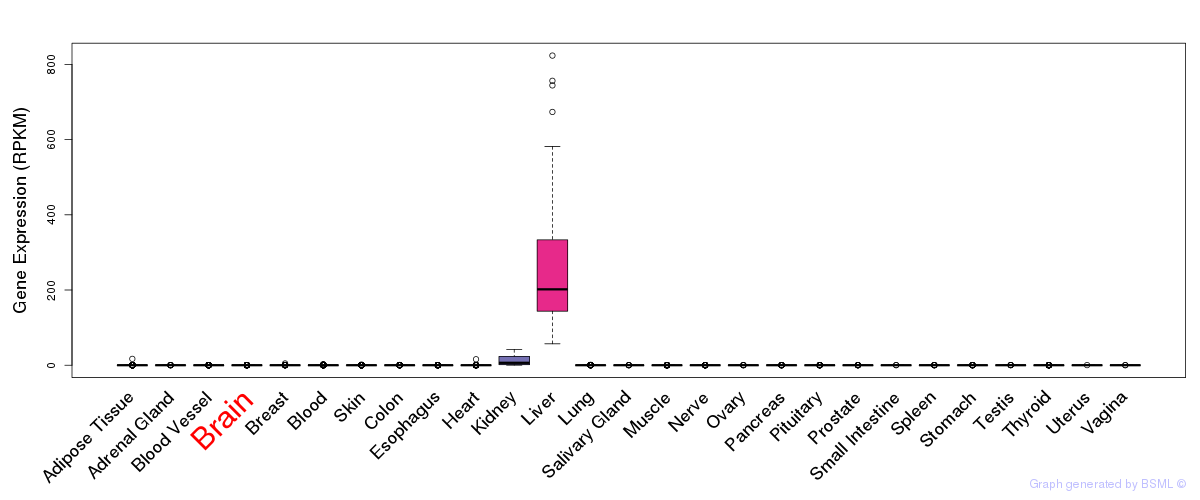
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| METTL11A | 0.90 | 0.89 |
| C19orf62 | 0.88 | 0.88 |
| AP2S1 | 0.88 | 0.87 |
| TIMM50 | 0.88 | 0.85 |
| MANBAL | 0.87 | 0.84 |
| CDK5 | 0.87 | 0.84 |
| TSFM | 0.87 | 0.82 |
| FIBP | 0.86 | 0.85 |
| ASNA1 | 0.86 | 0.83 |
| FLOT1 | 0.85 | 0.84 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.18 | -0.58 | -0.43 |
| AF347015.26 | -0.56 | -0.41 |
| AF347015.2 | -0.54 | -0.37 |
| AF347015.8 | -0.49 | -0.34 |
| AF347015.15 | -0.48 | -0.34 |
| MT-CO2 | -0.47 | -0.30 |
| MT-CYB | -0.47 | -0.33 |
| AC010300.1 | -0.47 | -0.47 |
| AC016705.1 | -0.47 | -0.40 |
| AL139819.3 | -0.47 | -0.39 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004252 | serine-type endopeptidase activity | TAS | glutamate (GO term level: 7) | 9242524 |
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0008233 | peptidase activity | IEA | - | |
| GO:0034185 | apolipoprotein binding | IPI | 16480936 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006508 | proteolysis | IEA | - | |
| GO:0006917 | induction of apoptosis | IEA | - | |
| GO:0006917 | induction of apoptosis | TAS | 11257124 | |
| GO:0008285 | negative regulation of cell proliferation | TAS | 8910613 | |
| GO:0007596 | blood coagulation | IMP | 1986355 | |
| GO:0006915 | apoptosis | IEA | - | |
| GO:0042246 | tissue regeneration | IEA | - | |
| GO:0016525 | negative regulation of angiogenesis | TAS | 11257124 | |
| GO:0043537 | negative regulation of blood vessel endothelial cell migration | TAS | 11257124 | |
| GO:0046716 | muscle maintenance | IEA | - | |
| GO:0045445 | myoblast differentiation | IEA | - | |
| GO:0048771 | tissue remodeling | IEA | - | |
| GO:0051918 | negative regulation of fibrinolysis | IDA | 14726399 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | EXP | 2318848 |2962641 | |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005615 | extracellular space | IDA | 6216475 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTG1 | ACT | ACTG | DFNA20 | DFNA26 | actin, gamma 1 | PLG interacts with ACTG1 (G-actin). This interaction was modelled on a demonstrated interaction between human PLG and bovine ACTG1. | BIND | 15746964 |
| AMOT | KIAA1071 | angiomotin | - | HPRD | 11257124 |
| ANXA2 | ANX2 | ANX2L4 | CAL1H | LIP2 | LPC2 | LPC2D | P36 | PAP-IV | annexin A2 | - | HPRD | 10077593 |
| ATP5B | ATPMB | ATPSB | MGC5231 | ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide | - | HPRD | 10077593 |
| C7 | - | complement component 7 | - | HPRD,BioGRID | 7814888 |
| CHGA | CGA | chromogranin A (parathyroid secretory protein 1) | - | HPRD,BioGRID | 11342539 |
| CLEC3B | DKFZp686H17246 | TN | TNA | C-type lectin domain family 3, member B | - | HPRD | 10964919 |
| CPB2 | CPU | PCPB | TAFI | carboxypeptidase B2 (plasma) | - | HPRD,BioGRID | 1939207 |
| CXCL2 | CINC-2a | GRO2 | GROb | MGSA-b | MIP-2a | MIP2 | MIP2A | SCYB2 | chemokine (C-X-C motif) ligand 2 | - | HPRD,BioGRID | 10928473 |
| ENO1 | ENO1L1 | MBP-1 | MPB1 | NNE | PPH | enolase 1, (alpha) | Reconstituted Complex | BioGRID | 11169399 |
| F10 | FX | FXA | coagulation factor X | - | HPRD,BioGRID | 11371191 |
| F3 | CD142 | TF | TFA | coagulation factor III (thromboplastin, tissue factor) | - | HPRD,BioGRID | 9490681 |
| HMGB1 | DKFZp686A04236 | HMG1 | HMG3 | SBP-1 | high-mobility group box 1 | - | HPRD,BioGRID | 1909331 |
| HRG | DKFZp779H1622 | HPRG | HRGP | histidine-rich glycoprotein | - | HPRD | 8478593 |
| IGFBP3 | BP-53 | IBP3 | insulin-like growth factor binding protein 3 | - | HPRD,BioGRID | 9688635 |
| KNG1 | BDK | KNG | kininogen 1 | - | HPRD,BioGRID | 9428707 |
| LAMA1 | LAMA | laminin, alpha 1 | - | HPRD,BioGRID | 8360181 |
| LAMA3 | BM600 | E170 | LAMNA | LOCS | lama3a | laminin, alpha 3 | - | HPRD | 10956663 |
| LAMA5 | KIAA1907 | laminin, alpha 5 | Reconstituted Complex | BioGRID | 10956663 |
| NGF | Beta-NGF | HSAN5 | MGC161426 | MGC161428 | NGFB | nerve growth factor (beta polypeptide) | - | HPRD | 11729324 |
| PAPPA | ASBABP2 | DIPLA1 | IGFBP-4ase | PAPA | PAPP-A | PAPPA1 | pregnancy-associated plasma protein A, pappalysin 1 | Biochemical Activity | BioGRID | 7586586 |
| PLAU | ATF | UPA | URK | UROKINASE | u-PA | plasminogen activator, urokinase | - | HPRD | 2829380 |3920216 |
| PRNP | ASCR | CD230 | CJD | GSS | MGC26679 | PRIP | PrP | PrP27-30 | PrP33-35C | PrPc | prion | prion protein | PrPSc interacts with PLG. | BIND | 11438139 |
| S100A10 | 42C | ANX2L | ANX2LG | CAL1L | CLP11 | Ca[1] | GP11 | MGC111133 | P11 | p10 | S100 calcium binding protein A10 | - | HPRD,BioGRID | 11939791 |
| SERPINC1 | AT3 | ATIII | MGC22579 | serpin peptidase inhibitor, clade C (antithrombin), member 1 | - | HPRD | 10974350 |
| SERPINE1 | PAI | PAI-1 | PAI1 | PLANH1 | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 | Reconstituted Complex | BioGRID | 11113116 |
| SERPINF2 | A2AP | AAP | ALPHA-2-PI | API | PLI | serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2 | - | HPRD,BioGRID | 158022 |2437112 |3334852 |
| SPARC | ON | secreted protein, acidic, cysteine-rich (osteonectin) | - | HPRD,BioGRID | 2745554 |7982919 |
| TFPI2 | FLJ21164 | PP5 | REF1 | TFPI-2 | tissue factor pathway inhibitor 2 | - | HPRD | 8555184 |
| THBS1 | THBS | TSP | TSP1 | thrombospondin 1 | - | HPRD,BioGRID | 2522013 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| KEGG COMPLEMENT AND COAGULATION CASCADES | 69 | 49 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AMI PATHWAY | 20 | 13 | All SZGR 2.0 genes in this pathway |
| BIOCARTA FIBRINOLYSIS PATHWAY | 12 | 8 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PLATELETAPP PATHWAY | 14 | 10 | All SZGR 2.0 genes in this pathway |
| PID ANGIOPOIETIN RECEPTOR PATHWAY | 50 | 41 | All SZGR 2.0 genes in this pathway |
| PID P75 NTR PATHWAY | 69 | 51 | All SZGR 2.0 genes in this pathway |
| PID AMB2 NEUTROPHILS PATHWAY | 41 | 32 | All SZGR 2.0 genes in this pathway |
| PID SYNDECAN 4 PATHWAY | 32 | 25 | All SZGR 2.0 genes in this pathway |
| PID UPA UPAR PATHWAY | 42 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | 29 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME EXTRACELLULAR MATRIX ORGANIZATION | 87 | 42 | All SZGR 2.0 genes in this pathway |
| REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | 89 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME DIABETES PATHWAYS | 133 | 91 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | 16 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY PDGF | 122 | 93 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| HUMMERICH SKIN CANCER PROGRESSION UP | 88 | 58 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN | 514 | 319 | All SZGR 2.0 genes in this pathway |
| RANKIN ANGIOGENIC TARGETS OF VHL HIF2A UP | 5 | 5 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| BROWN MYELOID CELL DEVELOPMENT UP | 165 | 100 | All SZGR 2.0 genes in this pathway |
| HSIAO LIVER SPECIFIC GENES | 244 | 154 | All SZGR 2.0 genes in this pathway |
| HOEGERKORP CD44 TARGETS DIRECT UP | 27 | 21 | All SZGR 2.0 genes in this pathway |
| XU GH1 EXOGENOUS TARGETS DN | 120 | 69 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER | 49 | 29 | All SZGR 2.0 genes in this pathway |
| OUILLETTE CLL 13Q14 DELETION DN | 60 | 38 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE DN | 274 | 165 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| LABBE TGFB1 TARGETS UP | 102 | 64 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS DN | 138 | 92 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE UP | 212 | 128 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS PROLIFERATION DN | 179 | 97 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS UNANNOTATED UP | 85 | 50 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SURVIVAL DN | 113 | 76 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S3 | 266 | 180 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT DN | 222 | 141 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| SERVITJA LIVER HNF1A TARGETS DN | 157 | 105 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
| NABA ECM REGULATORS | 238 | 125 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |