Gene Page: ATP7B
Summary ?
| GeneID | 540 |
| Symbol | ATP7B |
| Synonyms | PWD|WC1|WD|WND |
| Description | ATPase copper transporting beta |
| Reference | MIM:606882|HGNC:HGNC:870|Ensembl:ENSG00000123191|HPRD:06050|Vega:OTTHUMG00000017406 |
| Gene type | protein-coding |
| Map location | 13q14.3 |
| Pascal p-value | 0.402 |
| Sherlock p-value | 0.686 |
| Fetal beta | 0.073 |
| eGene | Cerebellar Hemisphere Cerebellum Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs461218 | chr5 | 73410191 | ATP7B | 540 | 0.19 | trans | ||
| rs2937774 | chr5 | 73420816 | ATP7B | 540 | 0.07 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
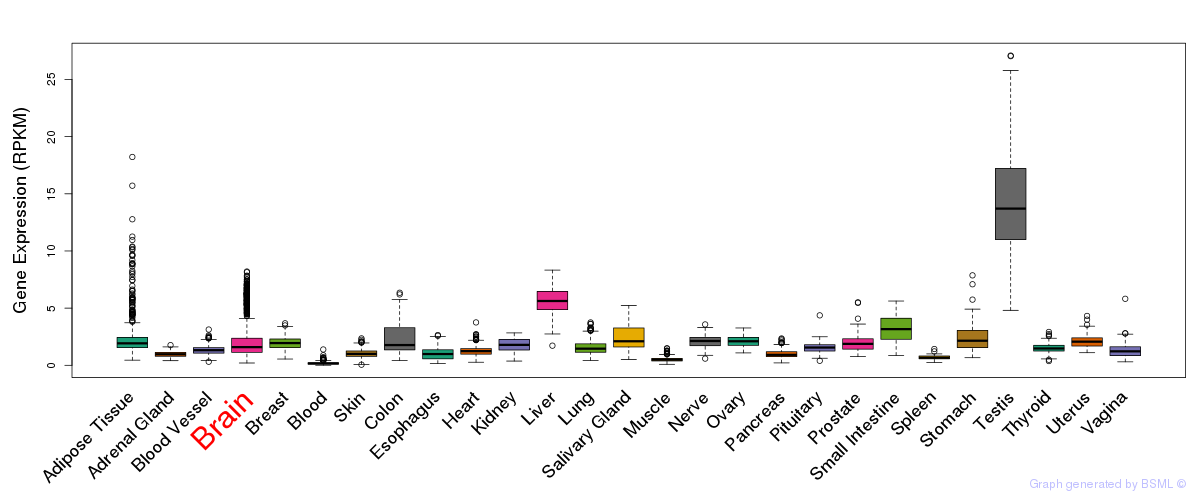
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0000287 | magnesium ion binding | IEA | - | |
| GO:0004008 | copper-exporting ATPase activity | IEA | - | |
| GO:0004008 | copper-exporting ATPase activity | NAS | 12763797 | |
| GO:0004008 | copper-exporting ATPase activity | TAS | 16472602 | |
| GO:0005375 | copper ion transmembrane transporter activity | IEA | - | |
| GO:0005507 | copper ion binding | IDA | 12029094 | |
| GO:0005507 | copper ion binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 16554302 |16884690 | |
| GO:0005524 | ATP binding | IDA | 16567646 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0016787 | hydrolase activity | IEA | - | |
| GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances | IEA | - | |
| GO:0015662 | ATPase activity, coupled to transmembrane movement of ions, phosphorylative mechanism | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| GO:0046873 | metal ion transmembrane transporter activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0030001 | metal ion transport | IEA | - | |
| GO:0008152 | metabolic process | IEA | - | |
| GO:0007595 | lactation | IEA | - | |
| GO:0006825 | copper ion transport | IEA | - | |
| GO:0006811 | ion transport | IEA | - | |
| GO:0006810 | transport | IEA | - | |
| GO:0006878 | cellular copper ion homeostasis | TAS | 16554302 | |
| GO:0006882 | cellular zinc ion homeostasis | IEA | - | |
| GO:0015680 | intracellular copper ion transport | IEA | - | |
| GO:0015677 | copper ion import | IDA | 16472602 | |
| GO:0051208 | sequestering of calcium ion | IDA | 16472602 | |
| GO:0046688 | response to copper ion | IDA | 15269005 |16472602 |16939419 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005794 | Golgi apparatus | IEA | - | |
| GO:0005802 | trans-Golgi network | IDA | 15269005 |16472602 | |
| GO:0005624 | membrane fraction | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005739 | mitochondrion | IEA | - | |
| GO:0005770 | late endosome | IDA | 15681833 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0016023 | cytoplasmic membrane-bounded vesicle | IDA | 16472602 | |
| GO:0005887 | integral to plasma membrane | TAS | 8298641 | |
| GO:0048471 | perinuclear region of cytoplasm | IDA | 16939419 | |
| GO:0016323 | basolateral plasma membrane | IDA | 15269005 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | 413 | 270 | All SZGR 2.0 genes in this pathway |
| REACTOME ION TRANSPORT BY P TYPE ATPASES | 34 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME ION CHANNEL TRANSPORT | 55 | 42 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| LAIHO COLORECTAL CANCER SERRATED DN | 86 | 47 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 8D UP | 157 | 91 | All SZGR 2.0 genes in this pathway |
| LUCAS HNF4A TARGETS UP | 58 | 36 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| GARCIA TARGETS OF FLI1 AND DAX1 UP | 57 | 34 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 16HR UP | 225 | 139 | All SZGR 2.0 genes in this pathway |
| JOSEPH RESPONSE TO SODIUM BUTYRATE DN | 64 | 45 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION DN | 287 | 208 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME3 AND H3K27ME3 | 142 | 103 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL SCP2 QTL TRANS | 25 | 13 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K4ME3 AND H3K27ME3 | 210 | 148 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS DN | 242 | 146 | All SZGR 2.0 genes in this pathway |