Gene Page: ADAMTSL4
Summary ?
| GeneID | 54507 |
| Symbol | ADAMTSL4 |
| Synonyms | ADAMTSL-4|ECTOL2|TSRC1 |
| Description | ADAMTS like 4 |
| Reference | MIM:610113|HGNC:HGNC:19706|Ensembl:ENSG00000143382|HPRD:08583|HPRD:18237|Vega:OTTHUMG00000034863 |
| Gene type | protein-coding |
| Map location | 1q21.3 |
| Fetal beta | -0.778 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0235 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00814 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
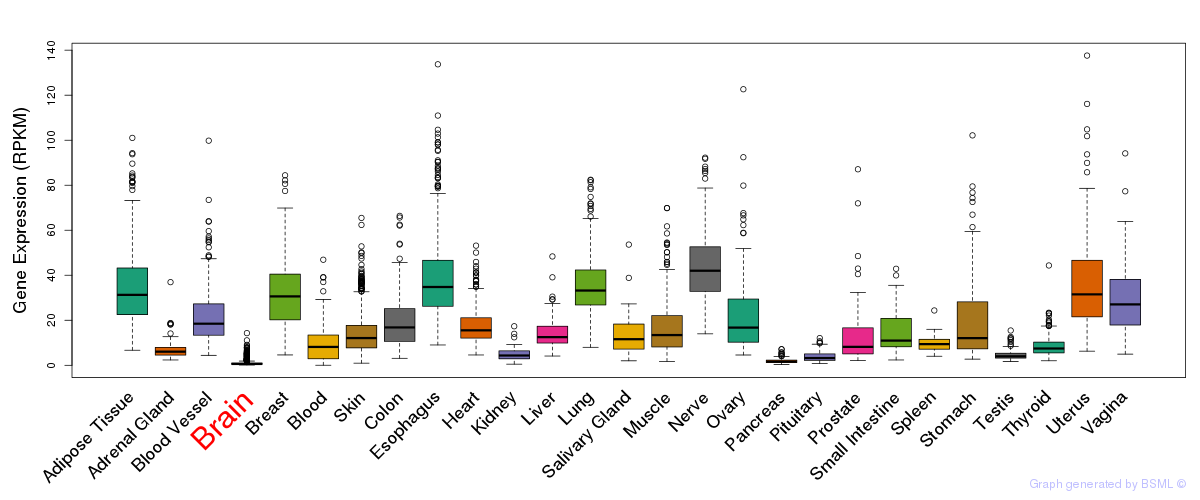
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0002020 | protease binding | IPI | 16364318 | |
| GO:0004222 | metalloendopeptidase activity | IEA | - | |
| GO:0008233 | peptidase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0043065 | positive regulation of apoptosis | IDA | 16364318 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005737 | cytoplasm | IDA | 18029348 | |
| GO:0005886 | plasma membrane | IDA | 18029348 | |
| GO:0031012 | extracellular matrix | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CATSPER1 | CATSPER | MGC33335 | MGC33368 | cation channel, sperm associated 1 | Two-hybrid | BioGRID | 16189514 |
| DLK2 | EGFL9 | MGC111055 | MGC2487 | delta-like 2 homolog (Drosophila) | Two-hybrid | BioGRID | 16189514 |
| DNPEP | ASPEP | DAP | aspartyl aminopeptidase | Two-hybrid | BioGRID | 16189514 |
| FHL3 | MGC19547 | MGC23614 | MGC8696 | SLIM2 | four and a half LIM domains 3 | Two-hybrid | BioGRID | 16189514 |
| GLRX3 | FLJ11864 | GLRX4 | GRX3 | GRX4 | PICOT | TXNL2 | TXNL3 | bA500G10.4 | glutaredoxin 3 | Two-hybrid | BioGRID | 16189514 |
| KRTAP4-12 | KAP4.12 | KRTAP4.12 | keratin associated protein 4-12 | Two-hybrid | BioGRID | 16189514 |
| NTN4 | FLJ23180 | PRO3091 | netrin 4 | Two-hybrid | BioGRID | 16189514 |
| PLSCR1 | MMTRA1B | phospholipid scramblase 1 | Two-hybrid | BioGRID | 16189514 |
| SORBS3 | SCAM-1 | SCAM1 | SH3D4 | VINEXIN | sorbin and SH3 domain containing 3 | Two-hybrid | BioGRID | 16189514 |
| SPRY2 | MGC23039 | hSPRY2 | sprouty homolog 2 (Drosophila) | Two-hybrid | BioGRID | 16189514 |
| STK16 | FLJ39635 | KRCT | MPSK | PKL12 | TSF1 | serine/threonine kinase 16 | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION ERYTHROCYTE UP | 157 | 104 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS UP | 149 | 84 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS F UP | 185 | 119 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP | 390 | 236 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS DN | 261 | 155 | All SZGR 2.0 genes in this pathway |
| GRADE COLON AND RECTAL CANCER DN | 101 | 65 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |
| ACOSTA PROLIFERATION INDEPENDENT MYC TARGETS DN | 116 | 74 | All SZGR 2.0 genes in this pathway |
| NABA ECM REGULATORS | 238 | 125 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |