Gene Page: PPARD
Summary ?
| GeneID | 5467 |
| Symbol | PPARD |
| Synonyms | FAAR|NR1C2|NUC1|NUCI|NUCII|PPARB |
| Description | peroxisome proliferator activated receptor delta |
| Reference | MIM:600409|HGNC:HGNC:9235|Ensembl:ENSG00000112033|HPRD:02679|Vega:OTTHUMG00000014567 |
| Gene type | protein-coding |
| Map location | 6p21.2 |
| Pascal p-value | 0.17 |
| Sherlock p-value | 0.032 |
| Fetal beta | -0.728 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.033 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.04433 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg02943769 | 6 | 35310542 | PPARD | 2.86E-6 | -0.289 | 0.009 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
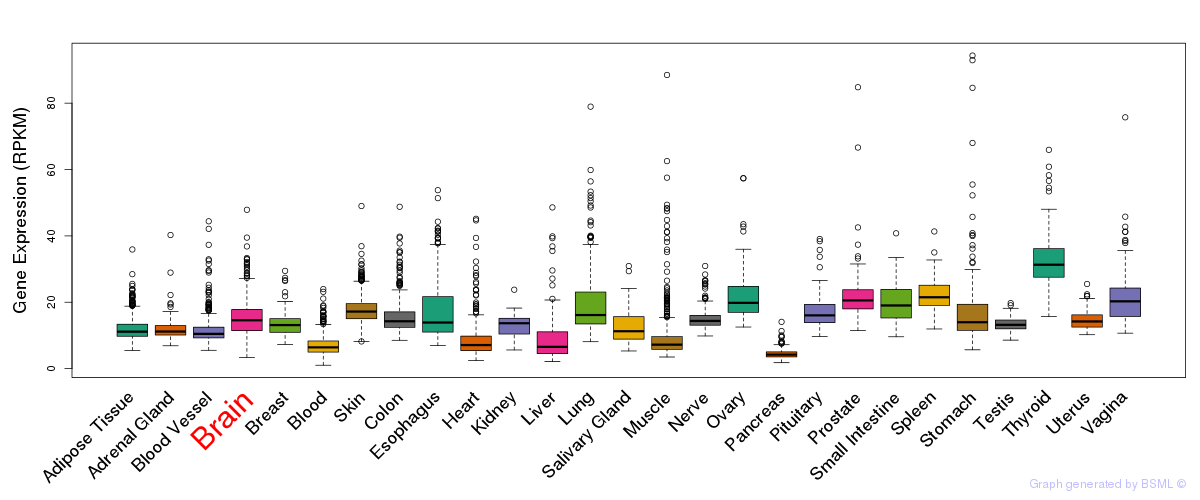
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MRPL38 | 0.88 | 0.84 |
| ERI3 | 0.88 | 0.85 |
| MRPL4 | 0.88 | 0.83 |
| MRPL17 | 0.88 | 0.84 |
| MRPS12 | 0.86 | 0.83 |
| MRPS7 | 0.86 | 0.83 |
| ARF5 | 0.85 | 0.82 |
| SLC25A11 | 0.85 | 0.83 |
| MRPS11 | 0.85 | 0.80 |
| MRP63 | 0.85 | 0.80 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.18 | -0.54 | -0.45 |
| AC010300.1 | -0.50 | -0.55 |
| AF347015.26 | -0.48 | -0.38 |
| MT-ATP8 | -0.46 | -0.38 |
| AF347015.2 | -0.46 | -0.35 |
| AF347015.8 | -0.46 | -0.36 |
| AC100783.1 | -0.45 | -0.40 |
| AF347015.15 | -0.44 | -0.34 |
| AC016705.1 | -0.43 | -0.43 |
| NSBP1 | -0.41 | -0.41 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | NAS | 11551955 | |
| GO:0003707 | steroid hormone receptor activity | TAS | 1333051 | |
| GO:0005504 | fatty acid binding | NAS | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0008144 | drug binding | IDA | 9113987 | |
| GO:0016564 | transcription repressor activity | ISS | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| GO:0043565 | sequence-specific DNA binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008366 | axon ensheathment | ISS | neuron, axon (GO term level: 12) | - |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | ISS | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0006091 | generation of precursor metabolites and energy | TAS | 1333051 | |
| GO:0006006 | glucose metabolic process | NAS | 15793256 | |
| GO:0008203 | cholesterol metabolic process | TAS | 15192438 | |
| GO:0008283 | cell proliferation | ISS | - | |
| GO:0008283 | cell proliferation | TAS | 11847022 | |
| GO:0008544 | epidermis development | ISS | - | |
| GO:0007566 | embryo implantation | TAS | 11551955 | |
| GO:0015758 | glucose transport | NAS | 15793256 | |
| GO:0006635 | fatty acid beta-oxidation | ISS | - | |
| GO:0006635 | fatty acid beta-oxidation | TAS | 15192438 |15803109 | |
| GO:0006629 | lipid metabolic process | ISS | - | |
| GO:0006915 | apoptosis | IMP | 11551955 | |
| GO:0014068 | positive regulation of phosphoinositide 3-kinase cascade | IEA | - | |
| GO:0015908 | fatty acid transport | ISS | - | |
| GO:0030154 | cell differentiation | IEA | - | |
| GO:0042640 | anagen | IEA | - | |
| GO:0045600 | positive regulation of fat cell differentiation | NAS | 10991946 | |
| GO:0046697 | decidualization | TAS | 11551955 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | NAS | 11551955 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BCL6 | BCL5 | BCL6A | LAZ3 | ZBTB27 | ZNF51 | B-cell CLL/lymphoma 6 | PPAR-delta interacts with BCL-6. This interaction was modelled on a demonstrated interaction between mouse PPAR-delta and human BCL-6. | BIND | 12970571 |
| CEP350 | CAP350 | FLJ38282 | FLJ44058 | GM133 | KIAA0480 | centrosomal protein 350kDa | CAP350 interacts with PPAR-delta. | BIND | 15615782 |
| DUT | FLJ20622 | dUTPase | deoxyuridine triphosphatase | - | HPRD | 8910358 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | - | HPRD | 10567538 |
| GADD45B | DKFZp566B133 | GADD45BETA | MYD118 | growth arrest and DNA-damage-inducible, beta | - | HPRD | 10872826 |
| GADD45G | CR6 | DDIT2 | GADD45gamma | GRP17 | growth arrest and DNA-damage-inducible, gamma | - | HPRD,BioGRID | 10872826 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | - | HPRD,BioGRID | 11867749 |
| HDAC2 | RPD3 | YAF1 | histone deacetylase 2 | - | HPRD,BioGRID | 11867749 |
| HDAC3 | HD3 | RPD3 | RPD3-2 | histone deacetylase 3 | Affinity Capture-Western Reconstituted Complex | BioGRID | 11867749 |12943985 |
| HDAC4 | HA6116 | HD4 | HDAC-A | HDACA | KIAA0288 | histone deacetylase 4 | Reconstituted Complex | BioGRID | 12943985 |
| HDAC7 | DKFZp586J0917 | FLJ99588 | HD7A | HDAC7A | histone deacetylase 7 | PPAR-delta interacts with HDAC7. This interaction was modelled on a demonstrated interaction between PPAR-delta and HDAC7 from unspecified sources. | BIND | 12970571 |
| NCOR1 | KIAA1047 | MGC104216 | N-CoR | TRAC1 | hCIT529I10 | hN-CoR | nuclear receptor co-repressor 1 | Two-hybrid | BioGRID | 11867749 |
| NCOR2 | CTG26 | SMRT | SMRTE | SMRTE-tau | TNRC14 | TRAC-1 | TRAC1 | nuclear receptor co-repressor 2 | - | HPRD,BioGRID | 11867749 |
| NCOR2 | CTG26 | SMRT | SMRTE | SMRTE-tau | TNRC14 | TRAC-1 | TRAC1 | nuclear receptor co-repressor 2 | PPAR-delta interacts with SMRT. This interaction was modelled on a demonstrated interaction between PPAR-delta and SMRT from unspecified sources. | BIND | 12970571 |
| SPEN | KIAA0929 | MINT | RP1-134O19.1 | SHARP | spen homolog, transcriptional regulator (Drosophila) | - | HPRD,BioGRID | 11867749 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PPAR SIGNALING PATHWAY | 69 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG WNT SIGNALING PATHWAY | 151 | 112 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG ACUTE MYELOID LEUKEMIA | 60 | 47 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NUCLEARRS PATHWAY | 15 | 12 | All SZGR 2.0 genes in this pathway |
| BIOCARTA WNT PATHWAY | 26 | 24 | All SZGR 2.0 genes in this pathway |
| PID PS1 PATHWAY | 46 | 39 | All SZGR 2.0 genes in this pathway |
| PID RXR VDR PATHWAY | 26 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERIC TRANSCRIPTION PATHWAY | 352 | 181 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | 49 | 36 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA UP | 92 | 57 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP | 579 | 346 | All SZGR 2.0 genes in this pathway |
| DOANE RESPONSE TO ANDROGEN DN | 241 | 146 | All SZGR 2.0 genes in this pathway |
| SMIRNOV CIRCULATING ENDOTHELIOCYTES IN CANCER UP | 158 | 103 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER 20Q13 AMPLIFICATION DN | 180 | 101 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 UP | 380 | 236 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION ERYTHROCYTE UP | 157 | 104 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS UP | 293 | 179 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY DN | 382 | 224 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS MODERATELY DN | 110 | 64 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO IKK INHIBITOR AND TNF UP | 223 | 140 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA DN | 289 | 166 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION MUSCLE UP | 43 | 33 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION DN | 287 | 208 | All SZGR 2.0 genes in this pathway |
| URS ADIPOCYTE DIFFERENTIATION DN | 30 | 20 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS UP | 317 | 208 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| LEE METASTASIS AND ALTERNATIVE SPLICING DN | 45 | 31 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| LABBE WNT3A TARGETS DN | 97 | 53 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP | 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6 | 456 | 285 | All SZGR 2.0 genes in this pathway |
| FONTAINE FOLLICULAR THYROID ADENOMA UP | 75 | 43 | All SZGR 2.0 genes in this pathway |
| SENGUPTA EBNA1 ANTICORRELATED | 173 | 85 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 1HR DN | 222 | 147 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 1 DN | 48 | 30 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 3 DN | 42 | 24 | All SZGR 2.0 genes in this pathway |
| SERVITJA LIVER HNF1A TARGETS DN | 157 | 105 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-138 | 368 | 375 | 1A,m8 | hsa-miR-138brain | AGCUGGUGUUGUGAAUC |
| miR-29 | 1949 | 1955 | m8 | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| miR-9 | 518 | 524 | 1A | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.