Gene Page: DPP8
Summary ?
| GeneID | 54878 |
| Symbol | DPP8 |
| Synonyms | DP8|DPRP-1|DPRP1|MST097|MSTP097|MSTP135|MSTP141 |
| Description | dipeptidyl peptidase 8 |
| Reference | MIM:606819|HGNC:HGNC:16490|Ensembl:ENSG00000074603|HPRD:07076|Vega:OTTHUMG00000133150 |
| Gene type | protein-coding |
| Map location | 15q22 |
| Pascal p-value | 0.435 |
| Sherlock p-value | 0.032 |
| Fetal beta | -0.948 |
| eGene | Hippocampus |
| Support | CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
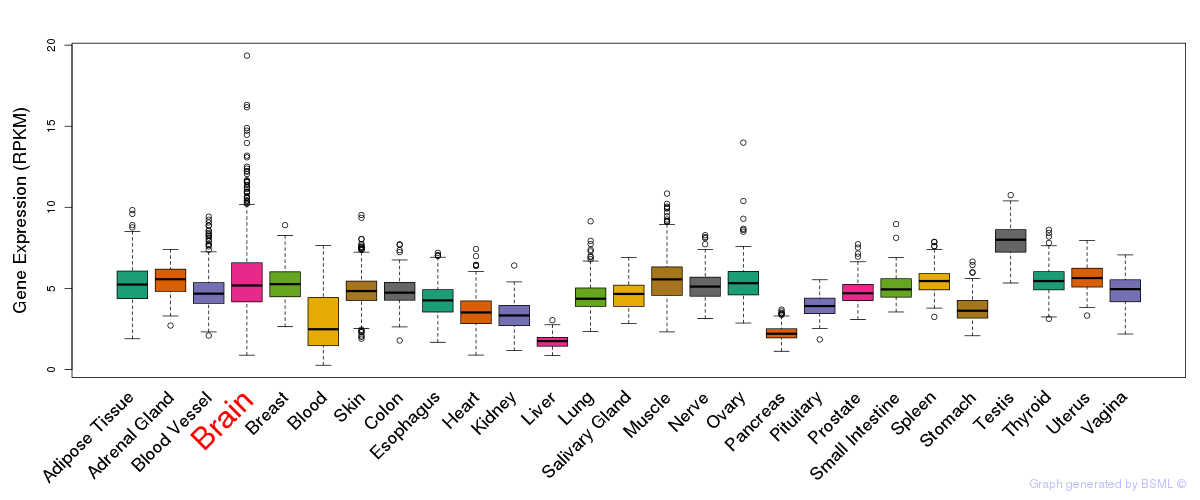
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RETSAT | 0.80 | 0.82 |
| C14orf159 | 0.80 | 0.83 |
| ITM2C | 0.79 | 0.80 |
| PAQR8 | 0.78 | 0.82 |
| SERPINE2 | 0.77 | 0.81 |
| DIO2 | 0.76 | 0.75 |
| TPP1 | 0.74 | 0.79 |
| NT5E | 0.74 | 0.80 |
| GNPTG | 0.74 | 0.79 |
| FAM167A | 0.74 | 0.74 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RBMX2 | -0.55 | -0.58 |
| CCAR1 | -0.53 | -0.53 |
| FRG1 | -0.51 | -0.50 |
| THOC2 | -0.50 | -0.47 |
| C11orf57 | -0.50 | -0.50 |
| EXOSC8 | -0.50 | -0.51 |
| PHF14 | -0.50 | -0.49 |
| RPL5 | -0.49 | -0.49 |
| ZNF300 | -0.49 | -0.35 |
| ZNF695 | -0.49 | -0.44 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004252 | serine-type endopeptidase activity | IEA | glutamate (GO term level: 7) | - |
| GO:0004177 | aminopeptidase activity | IEA | - | |
| GO:0008239 | dipeptidyl-peptidase activity | NAS | 11012666 | |
| GO:0008233 | peptidase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006508 | proteolysis | NAS | 11012666 | |
| GO:0006955 | immune response | TAS | 11012666 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IDA | 18029348 | |
| GO:0005737 | cytoplasm | NAS | 11012666 | |
| GO:0016020 | membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP | 633 | 376 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES SKIN UP | 177 | 113 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 DN | 229 | 142 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| DURCHDEWALD SKIN CARCINOGENESIS DN | 264 | 168 | All SZGR 2.0 genes in this pathway |
| CHEN HOXA5 TARGETS 9HR UP | 223 | 132 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS UP | 518 | 299 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-495 | 366 | 373 | 1A,m8 | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| miR-93.hd/291-3p/294/295/302/372/373/520 | 440 | 447 | 1A,m8 | hsa-miR-93brain | AAAGUGCUGUUCGUGCAGGUAG |
| hsa-miR-302a | UAAGUGCUUCCAUGUUUUGGUGA | ||||
| hsa-miR-302b | UAAGUGCUUCCAUGUUUUAGUAG | ||||
| hsa-miR-302c | UAAGUGCUUCCAUGUUUCAGUGG | ||||
| hsa-miR-302d | UAAGUGCUUCCAUGUUUGAGUGU | ||||
| hsa-miR-372 | AAAGUGCUGCGACAUUUGAGCGU | ||||
| hsa-miR-373 | GAAGUGCUUCGAUUUUGGGGUGU | ||||
| hsa-miR-520e | AAAGUGCUUCCUUUUUGAGGG | ||||
| hsa-miR-520a | AAAGUGCUUCCCUUUGGACUGU | ||||
| hsa-miR-520b | AAAGUGCUUCCUUUUAGAGGG | ||||
| hsa-miR-520c | AAAGUGCUUCCUUUUAGAGGGUU | ||||
| hsa-miR-520d | AAAGUGCUUCUCUUUGGUGGGUU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.