Gene Page: BANP
Summary ?
| GeneID | 54971 |
| Symbol | BANP |
| Synonyms | BEND1|SMAR1|SMARBP1 |
| Description | BTG3 associated nuclear protein |
| Reference | MIM:611564|HGNC:HGNC:13450|Ensembl:ENSG00000172530|HPRD:16538|Vega:OTTHUMG00000137678 |
| Gene type | protein-coding |
| Map location | 16q24 |
| Pascal p-value | 0.578 |
| Sherlock p-value | 0.503 |
| Fetal beta | 0.286 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| Expression | Meta-analysis of gene expression | P value: 1.453 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg03551062 | 16 | 88041289 | BANP | 2.943E-4 | -0.761 | 0.039 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16829545 | chr2 | 151977407 | BANP | 54971 | 0.01 | trans | ||
| rs7584986 | chr2 | 184111432 | BANP | 54971 | 0.04 | trans | ||
| snp_a-1815771 | 0 | BANP | 54971 | 0.03 | trans | |||
| rs10241465 | chr7 | 146018272 | BANP | 54971 | 0.1 | trans | ||
| rs3025425 | chr9 | 136523165 | BANP | 54971 | 0.1 | trans | ||
| rs16934084 | chr10 | 33430296 | BANP | 54971 | 0.11 | trans | ||
| rs7076439 | chr10 | 93286841 | BANP | 54971 | 0.01 | trans | ||
| rs11186601 | chr10 | 93287199 | BANP | 54971 | 0.01 | trans | ||
| rs2468302 | chr12 | 84899973 | BANP | 54971 | 0.19 | trans | ||
| rs16955618 | chr15 | 29937543 | BANP | 54971 | 0.05 | trans | ||
| rs16999433 | chr19 | 22772700 | BANP | 54971 | 0.09 | trans |
Section II. Transcriptome annotation
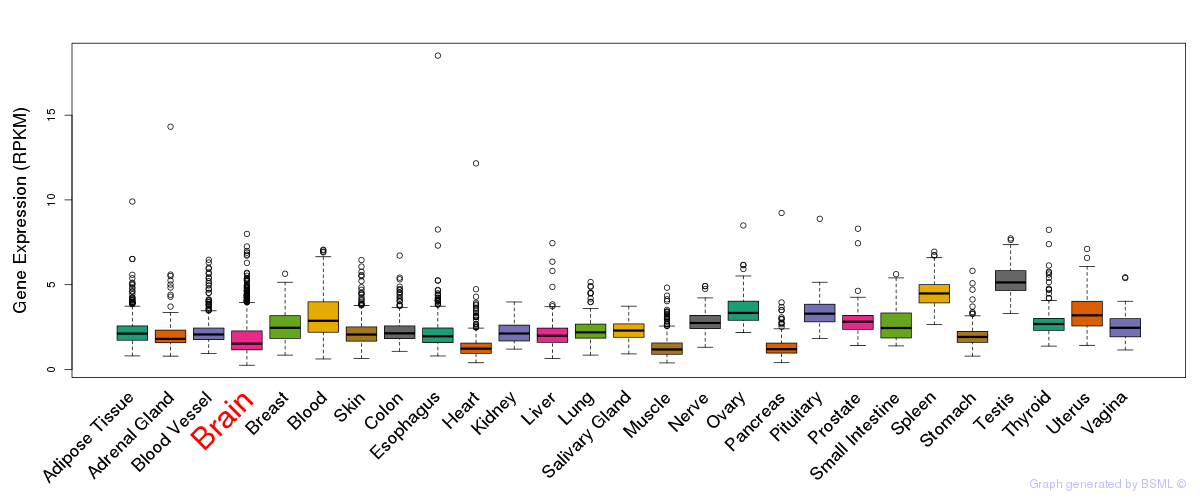
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| LDHD | 0.81 | 0.84 |
| RASSF4 | 0.81 | 0.79 |
| SEC14L2 | 0.80 | 0.80 |
| HSPB8 | 0.80 | 0.86 |
| DDIT3 | 0.79 | 0.82 |
| C5orf32 | 0.79 | 0.84 |
| TAPBPL | 0.79 | 0.80 |
| IRF9 | 0.78 | 0.85 |
| MX1 | 0.78 | 0.82 |
| GYG1 | 0.77 | 0.76 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KIAA1949 | -0.65 | -0.62 |
| PCDHB18 | -0.65 | -0.69 |
| SETDB1 | -0.65 | -0.65 |
| EPC1 | -0.65 | -0.67 |
| ZNF551 | -0.64 | -0.70 |
| AC004017.1 | -0.64 | -0.70 |
| TUBB2B | -0.64 | -0.62 |
| TOP2B | -0.64 | -0.69 |
| THOC2 | -0.63 | -0.73 |
| HNRNPAB | -0.63 | -0.67 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003677 | DNA binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0016568 | chromatin modification | IEA | - | |
| GO:0045786 | negative regulation of cell cycle | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| WANG CLIM2 TARGETS UP | 269 | 146 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL DN | 275 | 168 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS BLUE UP | 136 | 80 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP | 390 | 236 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 16Q24 AMPLICON | 53 | 38 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER BRACX DN | 20 | 14 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| SMIRNOV RESPONSE TO IR 2HR DN | 55 | 35 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-129-5p | 335 | 341 | 1A | hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC |
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| miR-320 | 303 | 309 | m8 | hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.