Gene Page: SLC47A1
Summary ?
| GeneID | 55244 |
| Symbol | SLC47A1 |
| Synonyms | MATE1 |
| Description | solute carrier family 47 member 1 |
| Reference | MIM:609832|HGNC:HGNC:25588|Ensembl:ENSG00000142494|HPRD:07703|Vega:OTTHUMG00000059466 |
| Gene type | protein-coding |
| Map location | 17p11.2 |
| Pascal p-value | 0.005 |
| Sherlock p-value | 0.805 |
| Fetal beta | -0.684 |
| DMG | 1 (# studies) |
| eGene | Nucleus accumbens basal ganglia Putamen basal ganglia Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg26959235 | 17 | 19437889 | SLC47A1 | 3.03E-4 | -0.302 | 0.04 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6587203 | chr17 | 19671457 | SLC47A1 | 55244 | 0.08 | cis | ||
| rs205113 | chr17 | 19673879 | SLC47A1 | 55244 | 0.08 | cis | ||
| rs205111 | chr17 | 19675103 | SLC47A1 | 55244 | 0.08 | cis | ||
| rs205110 | chr17 | 19675587 | SLC47A1 | 55244 | 0.08 | cis | ||
| rs205093 | chr17 | 19685712 | SLC47A1 | 55244 | 0.08 | cis | ||
| rs157406 | chr17 | 19710650 | SLC47A1 | 55244 | 0.08 | cis | ||
| rs157402 | chr17 | 19720588 | SLC47A1 | 55244 | 0.08 | cis | ||
| rs157396 | chr17 | 19726832 | SLC47A1 | 55244 | 0.08 | cis | ||
| rs157397 | chr17 | 19729494 | SLC47A1 | 55244 | 0.08 | cis | ||
| rs172186 | chr17 | 19780658 | SLC47A1 | 55244 | 0.03 | cis | ||
| rs281334 | chr17 | 19782079 | SLC47A1 | 55244 | 0.08 | cis | ||
| rs4586510 | chr17 | 19783973 | SLC47A1 | 55244 | 0.08 | cis | ||
| rs17131472 | chr1 | 91962966 | SLC47A1 | 55244 | 0.1 | trans | ||
| rs6718379 | chr2 | 18632808 | SLC47A1 | 55244 | 0.08 | trans | ||
| rs1016316 | chr2 | 20235108 | SLC47A1 | 55244 | 0.08 | trans | ||
| snp_a-2190074 | 0 | SLC47A1 | 55244 | 0.16 | trans | |||
| rs17535450 | chr2 | 39900012 | SLC47A1 | 55244 | 0.03 | trans | ||
| rs11706314 | chr3 | 45309716 | SLC47A1 | 55244 | 0 | trans | ||
| rs11706394 | chr3 | 45309907 | SLC47A1 | 55244 | 0.01 | trans | ||
| rs2922180 | chr3 | 125280472 | SLC47A1 | 55244 | 1.568E-5 | trans | ||
| rs2971271 | chr3 | 125450402 | SLC47A1 | 55244 | 0 | trans | ||
| rs2976809 | chr3 | 125451738 | SLC47A1 | 55244 | 4.074E-7 | trans | ||
| rs9917710 | chr3 | 176280218 | SLC47A1 | 55244 | 0.07 | trans | ||
| rs7668550 | chr4 | 104384355 | SLC47A1 | 55244 | 0 | trans | ||
| rs3796892 | chr4 | 111404774 | SLC47A1 | 55244 | 0.11 | trans | ||
| rs16894149 | chr5 | 61013724 | SLC47A1 | 55244 | 0.02 | trans | ||
| rs6449550 | chr5 | 61034312 | SLC47A1 | 55244 | 0.03 | trans | ||
| rs17242184 | chr5 | 71135066 | SLC47A1 | 55244 | 0.04 | trans | ||
| rs6874540 | chr5 | 149911669 | SLC47A1 | 55244 | 0.05 | trans | ||
| rs2273232 | chr5 | 149912526 | SLC47A1 | 55244 | 0.05 | trans | ||
| rs11954814 | chr5 | 155088338 | SLC47A1 | 55244 | 0.17 | trans | ||
| rs17056435 | chr5 | 158453782 | SLC47A1 | 55244 | 0.01 | trans | ||
| rs9313797 | chr5 | 158465457 | SLC47A1 | 55244 | 0.03 | trans | ||
| rs17056474 | chr5 | 158468105 | SLC47A1 | 55244 | 0.03 | trans | ||
| rs17135521 | chr6 | 2707631 | SLC47A1 | 55244 | 0.19 | trans | ||
| snp_a-1787926 | 0 | SLC47A1 | 55244 | 0.19 | trans | |||
| rs6939897 | chr6 | 67913123 | SLC47A1 | 55244 | 0.06 | trans | ||
| rs6455226 | chr6 | 67930295 | SLC47A1 | 55244 | 0 | trans | ||
| rs9387571 | chr6 | 118600137 | SLC47A1 | 55244 | 0.11 | trans | ||
| rs283058 | chr6 | 118625224 | SLC47A1 | 55244 | 0.01 | trans | ||
| rs9321275 | chr6 | 131493217 | SLC47A1 | 55244 | 0.03 | trans | ||
| rs2056744 | chr6 | 144952933 | SLC47A1 | 55244 | 0.12 | trans | ||
| rs17171399 | chr7 | 38592476 | SLC47A1 | 55244 | 0.08 | trans | ||
| rs2430483 | chr7 | 104548631 | SLC47A1 | 55244 | 0.01 | trans | ||
| rs2199402 | chr8 | 9201002 | SLC47A1 | 55244 | 0 | trans | ||
| rs2169384 | chr8 | 9216985 | SLC47A1 | 55244 | 0.1 | trans | ||
| rs1381350 | chr8 | 9217194 | SLC47A1 | 55244 | 0.14 | trans | ||
| rs7841407 | chr8 | 9243427 | SLC47A1 | 55244 | 8.51E-6 | trans | ||
| rs2739708 | chr8 | 17992280 | SLC47A1 | 55244 | 0.15 | trans | ||
| rs714131 | chr8 | 17992832 | SLC47A1 | 55244 | 0.15 | trans | ||
| rs2739726 | chr8 | 17998576 | SLC47A1 | 55244 | 0.08 | trans | ||
| rs4733365 | chr8 | 32480648 | SLC47A1 | 55244 | 0.01 | trans | ||
| rs2954041 | chr8 | 32522625 | SLC47A1 | 55244 | 0.11 | trans | ||
| rs2919377 | chr8 | 32573740 | SLC47A1 | 55244 | 0.01 | trans | ||
| rs2976526 | chr8 | 32574650 | SLC47A1 | 55244 | 0.11 | trans | ||
| rs2975504 | chr8 | 32578998 | SLC47A1 | 55244 | 0.11 | trans | ||
| rs17792062 | chr8 | 129163736 | SLC47A1 | 55244 | 0.15 | trans | ||
| rs7029518 | chr9 | 37782560 | SLC47A1 | 55244 | 0.12 | trans | ||
| rs427497 | chr9 | 107982791 | SLC47A1 | 55244 | 0.11 | trans | ||
| rs998410 | chr9 | 117622673 | SLC47A1 | 55244 | 0.08 | trans | ||
| rs9423711 | chr10 | 2574802 | SLC47A1 | 55244 | 0.01 | trans | ||
| rs748375 | chr10 | 33440178 | SLC47A1 | 55244 | 0.07 | trans | ||
| rs4514358 | chr10 | 43861923 | SLC47A1 | 55244 | 0.02 | trans | ||
| rs7081666 | chr10 | 47673800 | SLC47A1 | 55244 | 0.08 | trans | ||
| rs11820030 | chr11 | 76133564 | SLC47A1 | 55244 | 0.05 | trans | ||
| rs17134872 | chr11 | 76139845 | SLC47A1 | 55244 | 0.01 | trans | ||
| rs10751255 | 0 | SLC47A1 | 55244 | 0.07 | trans | |||
| rs11236774 | chr11 | 76234952 | SLC47A1 | 55244 | 0.01 | trans | ||
| rs4149623 | chr12 | 6450577 | SLC47A1 | 55244 | 0.14 | trans | ||
| rs11049036 | chr12 | 27650292 | SLC47A1 | 55244 | 0.08 | trans | ||
| rs10747972 | 0 | SLC47A1 | 55244 | 0.16 | trans | |||
| rs10879027 | chr12 | 70237156 | SLC47A1 | 55244 | 0.15 | trans | ||
| rs11112163 | chr12 | 105072508 | SLC47A1 | 55244 | 0.03 | trans | ||
| rs11067174 | chr12 | 115011927 | SLC47A1 | 55244 | 0.05 | trans | ||
| rs1750009 | chr13 | 42657093 | SLC47A1 | 55244 | 0 | trans | ||
| rs7995192 | chr13 | 50782598 | SLC47A1 | 55244 | 0.17 | trans | ||
| rs9596788 | chr13 | 53924430 | SLC47A1 | 55244 | 0.16 | trans | ||
| rs9529974 | chr13 | 72821935 | SLC47A1 | 55244 | 0 | trans | ||
| rs17720221 | chr13 | 75182377 | SLC47A1 | 55244 | 0.09 | trans | ||
| rs17073665 | chr13 | 81541961 | SLC47A1 | 55244 | 0.01 | trans | ||
| rs11840833 | chr13 | 94378306 | SLC47A1 | 55244 | 0.03 | trans | ||
| rs7319060 | chr13 | 95146303 | SLC47A1 | 55244 | 0.15 | trans | ||
| rs10142086 | chr14 | 35135892 | SLC47A1 | 55244 | 0.13 | trans | ||
| rs2093759 | chr14 | 97171024 | SLC47A1 | 55244 | 0.01 | trans | ||
| rs17244419 | chr14 | 97171074 | SLC47A1 | 55244 | 9.133E-4 | trans | ||
| rs7165622 | chr15 | 93979910 | SLC47A1 | 55244 | 0.1 | trans | ||
| rs13380712 | chr16 | 25601181 | SLC47A1 | 55244 | 0.02 | trans | ||
| rs17294918 | chr16 | 52717122 | SLC47A1 | 55244 | 0.08 | trans | ||
| rs17841422 | chr17 | 20969134 | SLC47A1 | 55244 | 0.03 | trans | ||
| rs8079075 | chr17 | 38010814 | SLC47A1 | 55244 | 0.18 | trans | ||
| rs16940550 | chr18 | 13442256 | SLC47A1 | 55244 | 0.19 | trans | ||
| rs7236407 | chr18 | 47827597 | SLC47A1 | 55244 | 0.08 | trans | ||
| rs4987765 | chr18 | 60905021 | SLC47A1 | 55244 | 0.17 | trans | ||
| rs11873703 | chr18 | 61448702 | SLC47A1 | 55244 | 5.818E-6 | trans | ||
| rs6095741 | chr20 | 48666589 | SLC47A1 | 55244 | 1.222E-5 | trans | ||
| rs2408239 | chr21 | 23332625 | SLC47A1 | 55244 | 0.13 | trans | ||
| rs17191809 | chr21 | 36444778 | SLC47A1 | 55244 | 0.04 | trans | ||
| rs6000401 | chr22 | 37149335 | SLC47A1 | 55244 | 0.05 | trans | ||
| rs5991662 | chrX | 43335796 | SLC47A1 | 55244 | 0.06 | trans | ||
| rs35416333 | 17 | 19427842 | SLC47A1 | ENSG00000142494.9 | 3.106E-7 | 0.01 | 29144 | gtex_brain_putamen_basal |
| rs2440164 | 17 | 19428610 | SLC47A1 | ENSG00000142494.9 | 7.362E-7 | 0.01 | 29912 | gtex_brain_putamen_basal |
| rs2440165 | 17 | 19428719 | SLC47A1 | ENSG00000142494.9 | 1.174E-6 | 0.01 | 30021 | gtex_brain_putamen_basal |
| rs11870808 | 17 | 19429379 | SLC47A1 | ENSG00000142494.9 | 9.186E-7 | 0.01 | 30681 | gtex_brain_putamen_basal |
| rs8071709 | 17 | 19429790 | SLC47A1 | ENSG00000142494.9 | 1.267E-6 | 0.01 | 31092 | gtex_brain_putamen_basal |
| rs2453578 | 17 | 19430645 | SLC47A1 | ENSG00000142494.9 | 1.269E-6 | 0.01 | 31947 | gtex_brain_putamen_basal |
| rs111374932 | 17 | 19431959 | SLC47A1 | ENSG00000142494.9 | 1.624E-6 | 0.01 | 33261 | gtex_brain_putamen_basal |
| rs11871125 | 17 | 19432393 | SLC47A1 | ENSG00000142494.9 | 1.066E-6 | 0.01 | 33695 | gtex_brain_putamen_basal |
| rs2453579 | 17 | 19435278 | SLC47A1 | ENSG00000142494.9 | 4.344E-6 | 0.01 | 36580 | gtex_brain_putamen_basal |
| rs2252281 | 17 | 19437187 | SLC47A1 | ENSG00000142494.9 | 6.098E-7 | 0.01 | 38489 | gtex_brain_putamen_basal |
| rs2453580 | 17 | 19438321 | SLC47A1 | ENSG00000142494.9 | 1.261E-6 | 0.01 | 39623 | gtex_brain_putamen_basal |
| rs2453581 | 17 | 19438560 | SLC47A1 | ENSG00000142494.9 | 2.583E-7 | 0.01 | 39862 | gtex_brain_putamen_basal |
| rs370142316 | 17 | 19438749 | SLC47A1 | ENSG00000142494.9 | 1.213E-6 | 0.01 | 40051 | gtex_brain_putamen_basal |
| rs2453582 | 17 | 19439066 | SLC47A1 | ENSG00000142494.9 | 2.583E-7 | 0.01 | 40368 | gtex_brain_putamen_basal |
| rs894680 | 17 | 19440538 | SLC47A1 | ENSG00000142494.9 | 1.768E-7 | 0.01 | 41840 | gtex_brain_putamen_basal |
| rs2453583 | 17 | 19442036 | SLC47A1 | ENSG00000142494.9 | 7.088E-7 | 0.01 | 43338 | gtex_brain_putamen_basal |
| rs2453584 | 17 | 19443445 | SLC47A1 | ENSG00000142494.9 | 2.198E-7 | 0.01 | 44747 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
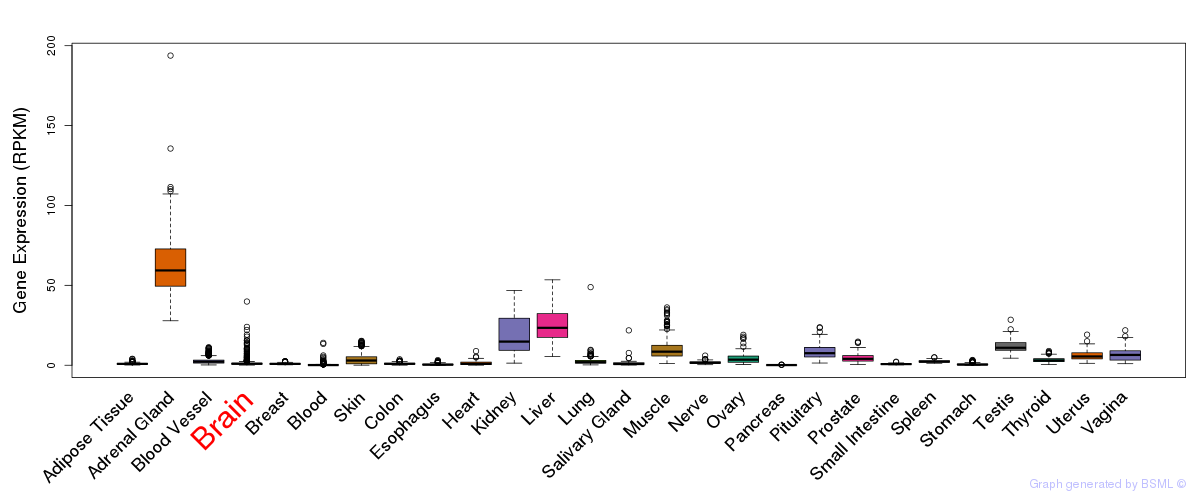
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | 413 | 270 | All SZGR 2.0 genes in this pathway |
| REACTOME SLC MEDIATED TRANSMEMBRANE TRANSPORT | 241 | 157 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS | 89 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | 11 | 6 | All SZGR 2.0 genes in this pathway |
| LIU PROSTATE CANCER DN | 481 | 290 | All SZGR 2.0 genes in this pathway |
| COLDREN GEFITINIB RESISTANCE UP | 85 | 57 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP | 390 | 236 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM4 | 261 | 153 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 17P11 AMPLICON | 10 | 8 | All SZGR 2.0 genes in this pathway |
| HELLER SILENCED BY METHYLATION DN | 105 | 67 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS DN | 292 | 189 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION DN | 281 | 179 | All SZGR 2.0 genes in this pathway |
| CLAUS PGR POSITIVE MENINGIOMA UP | 10 | 7 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA D UP | 89 | 62 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED | 536 | 296 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K27ME3 | 269 | 159 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS CTNNB1 UP | 176 | 110 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA PCA3 UP | 80 | 54 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 6HR UP | 229 | 149 | All SZGR 2.0 genes in this pathway |
| OHGUCHI LIVER HNF4A TARGETS DN | 149 | 85 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS NOT VIA AKT1 UP | 211 | 131 | All SZGR 2.0 genes in this pathway |
| SUMI HNF4A TARGETS | 34 | 14 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |