Gene Page: STAP2
Summary ?
| GeneID | 55620 |
| Symbol | STAP2 |
| Synonyms | BKS |
| Description | signal transducing adaptor family member 2 |
| Reference | MIM:607881|HGNC:HGNC:30430|Ensembl:ENSG00000178078|HPRD:07430|Vega:OTTHUMG00000181884 |
| Gene type | protein-coding |
| Map location | 19p13.3 |
| Pascal p-value | 0.799 |
| Sherlock p-value | 0.96 |
| Fetal beta | -0.898 |
| eGene | Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Frontal Cortex BA9 Hypothalamus Nucleus accumbens basal ganglia Putamen basal ganglia Myers' cis & trans Meta |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Xu_2012 | Whole Exome Sequencing analysis | De novo mutations of 4 genes were identified by exome sequencing of 795 samples in this study |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| STAP2 | chr19 | 4338685 | A | T | NM_001013841 NM_017720 | p.22Y>* p.22Y>* | nonsense nonsense | Schizophrenia | DNM:Xu_2012 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7258060 | chr19 | 4332104 | STAP2 | 55620 | 1.158E-4 | cis | ||
| rs7258060 | chr19 | 4332104 | STAP2 | 55620 | 0.01 | trans | ||
| rs374392257 | 19 | 4331940 | STAP2 | ENSG00000178078.7 | 1.29977E-6 | 0.01 | 10843 | gtex_brain_putamen_basal |
| rs7258060 | 19 | 4332105 | STAP2 | ENSG00000178078.7 | 5.24181E-7 | 0.01 | 10678 | gtex_brain_putamen_basal |
| rs11673317 | 19 | 4332650 | STAP2 | ENSG00000178078.7 | 2.64591E-7 | 0.01 | 10133 | gtex_brain_putamen_basal |
| rs10413905 | 19 | 4333666 | STAP2 | ENSG00000178078.7 | 5.06307E-7 | 0.01 | 9117 | gtex_brain_putamen_basal |
| rs60010631 | 19 | 4334310 | STAP2 | ENSG00000178078.7 | 5.00993E-7 | 0.01 | 8473 | gtex_brain_putamen_basal |
| rs55913120 | 19 | 4334402 | STAP2 | ENSG00000178078.7 | 1.56962E-6 | 0.01 | 8381 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
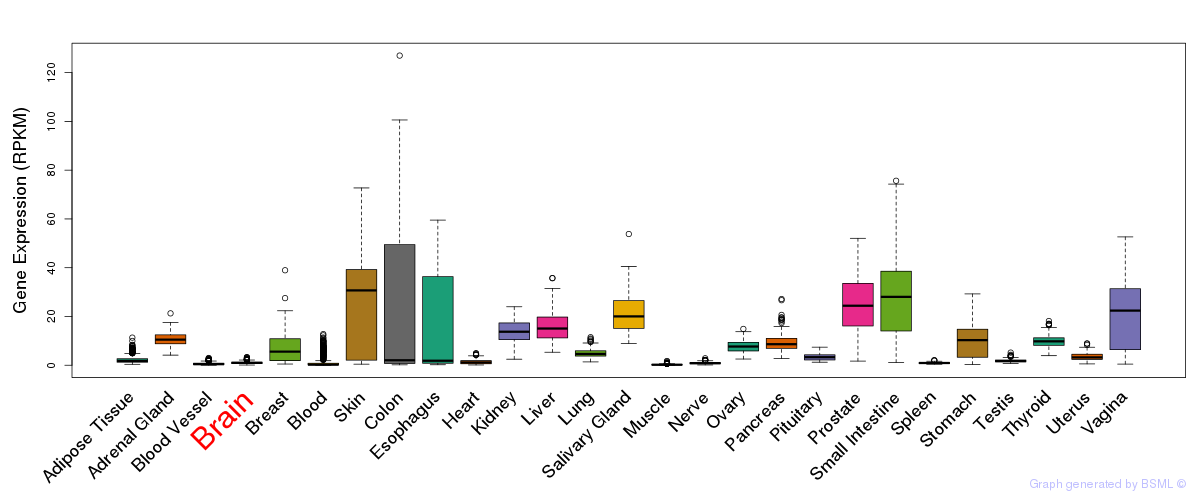
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GAB1 | 0.84 | 0.84 |
| TRIM59 | 0.80 | 0.79 |
| JAM3 | 0.80 | 0.84 |
| BBX | 0.79 | 0.84 |
| MYBL1 | 0.78 | 0.84 |
| TMTC2 | 0.77 | 0.81 |
| CCNE2 | 0.76 | 0.79 |
| SEPT10 | 0.76 | 0.71 |
| SLAIN1 | 0.75 | 0.66 |
| CREB5 | 0.74 | 0.84 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ST20 | -0.34 | -0.45 |
| TBC1D10A | -0.32 | -0.38 |
| PRR5 | -0.31 | -0.32 |
| SYCP3 | -0.31 | -0.37 |
| IL32 | -0.30 | -0.38 |
| RP9P | -0.30 | -0.46 |
| AC016757.1 | -0.30 | -0.35 |
| AC087071.1 | -0.30 | -0.43 |
| DUSP23 | -0.29 | -0.33 |
| EMID1 | -0.29 | -0.38 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| JAEGER METASTASIS DN | 258 | 141 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| COLDREN GEFITINIB RESISTANCE DN | 230 | 115 | All SZGR 2.0 genes in this pathway |
| SCHEIDEREIT IKK INTERACTING PROTEINS | 58 | 45 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN | 464 | 276 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS DN | 145 | 93 | All SZGR 2.0 genes in this pathway |
| GRADE METASTASIS DN | 45 | 31 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL DN | 428 | 246 | All SZGR 2.0 genes in this pathway |