Gene Page: UBAP2
Summary ?
| GeneID | 55833 |
| Symbol | UBAP2 |
| Synonyms | UBAP-2 |
| Description | ubiquitin associated protein 2 |
| Reference | HGNC:HGNC:14185|Ensembl:ENSG00000137073|HPRD:06748|Vega:OTTHUMG00000000427 |
| Gene type | protein-coding |
| Map location | 9p13.3 |
| Pascal p-value | 0.019 |
| Sherlock p-value | 0.389 |
| Fetal beta | 0.177 |
| DMG | 2 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 3 |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 3 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg21511069 | 9 | 34049294 | UBAP2 | -0.032 | 0.49 | DMG:Nishioka_2013 | |
| cg21511069 | 9 | 34049294 | UBAP2 | 1.98E-9 | -0.025 | 1.63E-6 | DMG:Jaffe_2016 |
| cg23029491 | 9 | 34048775 | UBAP2 | 2.67E-9 | -0.011 | 1.94E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
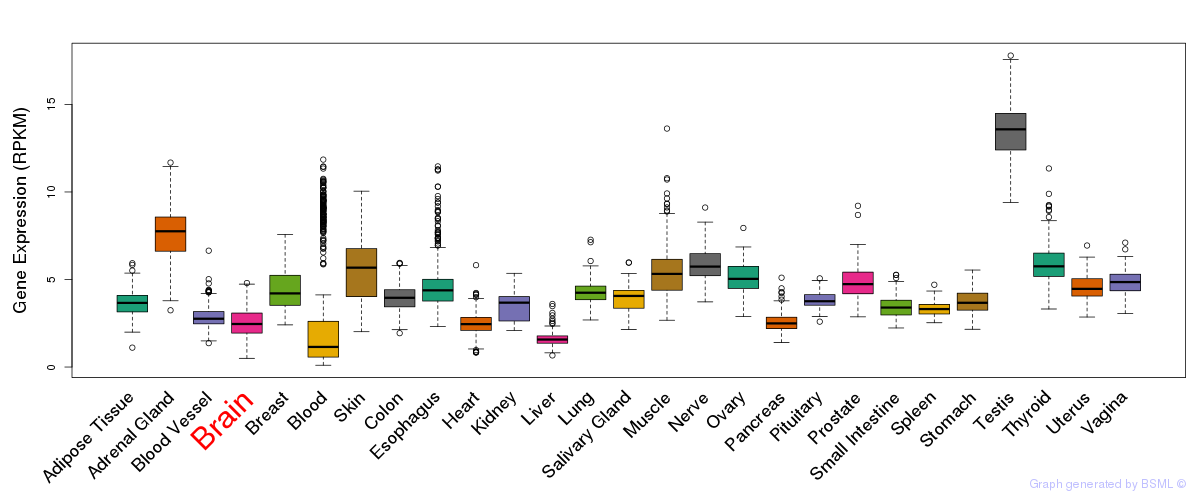
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ANKIB1 | 0.86 | 0.91 |
| AGL | 0.83 | 0.88 |
| USP34 | 0.82 | 0.87 |
| SLC11A2 | 0.82 | 0.88 |
| FAM120B | 0.81 | 0.88 |
| ZRANB1 | 0.81 | 0.86 |
| LPHN3 | 0.81 | 0.90 |
| PUM1 | 0.81 | 0.86 |
| RNF145 | 0.81 | 0.85 |
| CSMD3 | 0.81 | 0.84 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.67 | -0.73 |
| AF347015.31 | -0.66 | -0.71 |
| AF347015.21 | -0.65 | -0.75 |
| FXYD1 | -0.63 | -0.68 |
| AF347015.8 | -0.63 | -0.70 |
| IFI27 | -0.62 | -0.68 |
| AF347015.27 | -0.62 | -0.66 |
| HIGD1B | -0.61 | -0.70 |
| AF347015.33 | -0.61 | -0.64 |
| MT-CYB | -0.61 | -0.66 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| DITTMER PTHLH TARGETS DN | 73 | 51 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES CD4 DN | 116 | 71 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS G UP | 238 | 135 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN | 514 | 319 | All SZGR 2.0 genes in this pathway |
| LIAO HAVE SOX4 BINDING SITES | 40 | 26 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS | 539 | 324 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G1 UP | 113 | 70 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA UP | 207 | 143 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |