Gene Page: TDRD1
Summary ?
| GeneID | 56165 |
| Symbol | TDRD1 |
| Synonyms | CT41.1 |
| Description | tudor domain containing 1 |
| Reference | MIM:605796|HGNC:HGNC:11712|Ensembl:ENSG00000095627|HPRD:12047|Vega:OTTHUMG00000019083 |
| Gene type | protein-coding |
| Map location | 10q25.3 |
| Pascal p-value | 0.381 |
| Fetal beta | -0.337 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Gulsuner_2013 | Whole Exome Sequencing analysis | 155 DNMs identified by exome sequencing of quads or trios of schizophrenia individuals and their parents. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| TDRD1 | chr10 | 115973763 | A | G | NM_198795 | p.701N>S | missense | Schizophrenia | DNM:Gulsuner_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17016285 | chr1 | 106235493 | TDRD1 | 56165 | 0.09 | trans | ||
| rs16899821 | chr6 | 167451006 | TDRD1 | 56165 | 4.388E-4 | trans | ||
| rs16899825 | chr6 | 167451525 | TDRD1 | 56165 | 4.388E-4 | trans | ||
| rs9459881 | chr6 | 167524431 | TDRD1 | 56165 | 6.229E-4 | trans | ||
| rs11171021 | chr12 | 55083348 | TDRD1 | 56165 | 0.14 | trans | ||
| rs16965683 | chr15 | 38115683 | TDRD1 | 56165 | 0 | trans |
Section II. Transcriptome annotation
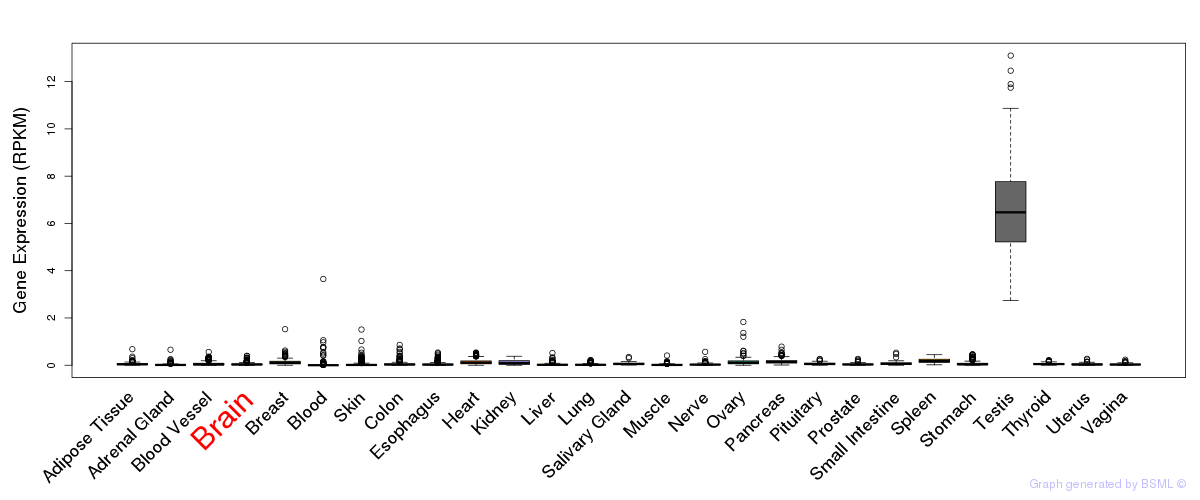
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| ODONNELL METASTASIS UP | 82 | 58 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS DN | 234 | 137 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR UP | 174 | 96 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| MATZUK SPERMATID DIFFERENTIATION | 37 | 26 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED | 536 | 296 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3 UNMETHYLATED | 37 | 21 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF ICP WITH H3K27ME3 | 206 | 108 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS WITH HCP H3K27ME3 | 102 | 76 | All SZGR 2.0 genes in this pathway |
| WEBER METHYLATED ICP IN FIBROBLAST | 22 | 9 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES HCP WITH H3K27ME3 | 41 | 30 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3 UNMETHYLATED | 228 | 119 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS DN | 414 | 237 | All SZGR 2.0 genes in this pathway |