Gene Page: CHST7
Summary ?
| GeneID | 56548 |
| Symbol | CHST7 |
| Synonyms | C6ST-2|GST-5 |
| Description | carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 |
| Reference | MIM:300375|HGNC:HGNC:13817|Ensembl:ENSG00000147119|HPRD:02302|Vega:OTTHUMG00000021423 |
| Gene type | protein-coding |
| Map location | Xp11.23 |
| Sherlock p-value | 0.89 |
| Fetal beta | -0.36 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs760697 | chrX | 46447343 | CHST7 | 56548 | 6.093E-6 | cis | ||
| rs5905551 | chrX | 46461535 | CHST7 | 56548 | 1.478E-4 | cis | ||
| rs5952927 | chrX | 46562924 | CHST7 | 56548 | 0.2 | cis | ||
| rs6980720 | chr8 | 127075270 | CHST7 | 56548 | 0.17 | trans | ||
| rs11015223 | chr10 | 26971331 | CHST7 | 56548 | 0.14 | trans | ||
| rs760697 | chrX | 46447343 | CHST7 | 56548 | 9.985E-4 | trans | ||
| rs5905551 | chrX | 46461535 | CHST7 | 56548 | 0.01 | trans |
Section II. Transcriptome annotation
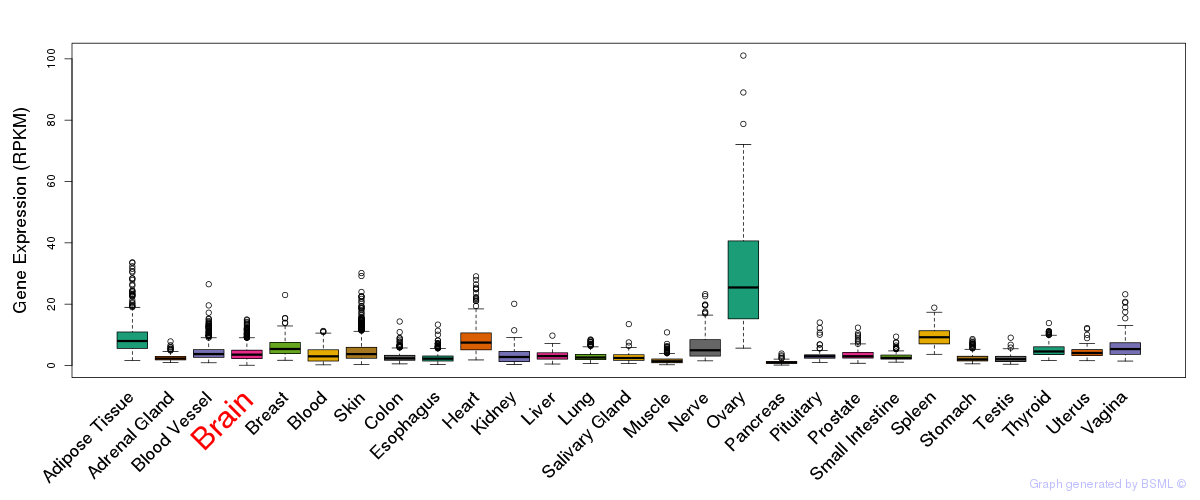
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ACD | 0.92 | 0.92 |
| TOR2A | 0.92 | 0.90 |
| LMBR1L | 0.92 | 0.92 |
| PRR14 | 0.91 | 0.88 |
| MIIP | 0.90 | 0.91 |
| FAM127B | 0.90 | 0.89 |
| PTOV1 | 0.90 | 0.90 |
| NARF | 0.90 | 0.89 |
| PCIF1 | 0.90 | 0.85 |
| C17orf62 | 0.89 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.27 | -0.69 | -0.82 |
| AF347015.31 | -0.67 | -0.78 |
| MT-CO2 | -0.67 | -0.78 |
| AF347015.8 | -0.67 | -0.81 |
| MT-CYB | -0.66 | -0.80 |
| AF347015.33 | -0.66 | -0.79 |
| AF347015.15 | -0.65 | -0.80 |
| ABCG2 | -0.64 | -0.70 |
| AIFM3 | -0.64 | -0.69 |
| AF347015.2 | -0.63 | -0.80 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity | IDA | 10913333 | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0008459 | chondroitin 6-sulfotransferase activity | IDA | 10781596 | |
| GO:0034482 | chondroitin 2-O-sulfotransferase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006044 | N-acetylglucosamine metabolic process | IDA | 10913333 | |
| GO:0005976 | polysaccharide metabolic process | TAS | 10781596 | |
| GO:0006790 | sulfur metabolic process | IDA | 10913333 | |
| GO:0030206 | chondroitin sulfate biosynthetic process | IDA | 10781596 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000139 | Golgi membrane | IEA | - | |
| GO:0005794 | Golgi apparatus | IEA | - | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | NAS | 10913333 | |
| GO:0016021 | integral to membrane | TAS | 10781596 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG GLYCOSAMINOGLYCAN BIOSYNTHESIS CHONDROITIN SULFATE | 22 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | 21 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | 49 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME GLYCOSAMINOGLYCAN METABOLISM | 111 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF CARBOHYDRATES | 247 | 154 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 1 DN | 126 | 86 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 2 DN | 77 | 46 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 DN | 162 | 116 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| PASQUALUCCI LYMPHOMA BY GC STAGE DN | 165 | 104 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES 1 | 379 | 235 | All SZGR 2.0 genes in this pathway |
| OKUMURA INFLAMMATORY RESPONSE LPS | 183 | 115 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| XU GH1 AUTOCRINE TARGETS UP | 268 | 157 | All SZGR 2.0 genes in this pathway |
| MAHAJAN RESPONSE TO IL1A UP | 81 | 52 | All SZGR 2.0 genes in this pathway |
| BILD E2F3 ONCOGENIC SIGNATURE | 246 | 153 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE DN | 274 | 165 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA D UP | 89 | 62 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA DN | 267 | 160 | All SZGR 2.0 genes in this pathway |
| HOELZEL NF1 TARGETS DN | 115 | 73 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS DN | 229 | 135 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-203.1 | 312 | 318 | 1A | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| miR-23 | 395 | 402 | 1A,m8 | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-323 | 395 | 401 | 1A | hsa-miR-323brain | GCACAUUACACGGUCGACCUCU |
| miR-328 | 405 | 412 | 1A,m8 | hsa-miR-328brain | CUGGCCCUCUCUGCCCUUCCGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.