Gene Page: PTK2
Summary ?
| GeneID | 5747 |
| Symbol | PTK2 |
| Synonyms | FADK|FAK|FAK1|FRNK|PPP1R71|p125FAK|pp125FAK |
| Description | protein tyrosine kinase 2 |
| Reference | MIM:600758|HGNC:HGNC:9611|Ensembl:ENSG00000169398|HPRD:02859|Vega:OTTHUMG00000067438 |
| Gene type | protein-coding |
| Map location | 8q24.3 |
| Pascal p-value | 0.04 |
| Sherlock p-value | 0.065 |
| Fetal beta | -0.566 |
| DMG | 1 (# studies) |
| eGene | Caudate basal ganglia Cerebellar Hemisphere Cerebellum |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 4 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 1.1049 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg03277652 | 8 | 142011117 | PTK2 | 1.89E-5 | -0.263 | 0.016 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
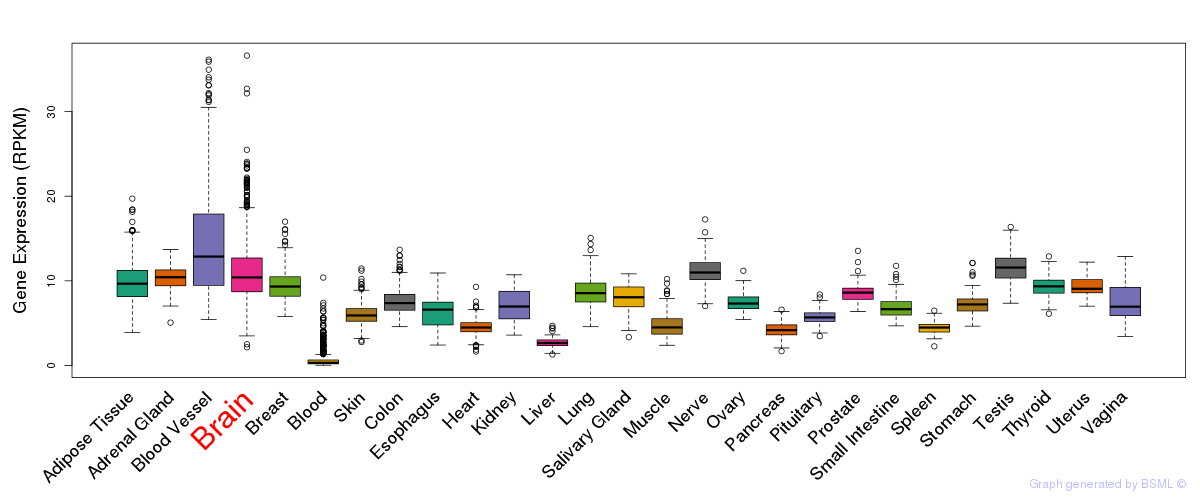
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0004871 | signal transducer activity | IEA | - | |
| GO:0005515 | protein binding | IPI | 8649368 |10822899 |14688263 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004713 | protein tyrosine kinase activity | IEA | - | |
| GO:0004715 | non-membrane spanning protein tyrosine kinase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0042169 | SH2 domain binding | IPI | 15077193 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001764 | neuron migration | IEA | neuron (GO term level: 8) | - |
| GO:0051964 | negative regulation of synaptogenesis | IEA | Synap (GO term level: 10) | - |
| GO:0021955 | central nervous system neuron axonogenesis | IEA | neuron, axon (GO term level: 13) | - |
| GO:0050771 | negative regulation of axonogenesis | IEA | axon, neurogenesis (GO term level: 14) | - |
| GO:0000226 | microtubule cytoskeleton organization | IEA | - | |
| GO:0001570 | vasculogenesis | IEA | - | |
| GO:0001568 | blood vessel development | IEA | - | |
| GO:0001525 | angiogenesis | IEA | - | |
| GO:0007172 | signal complex assembly | IEA | - | |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0007229 | integrin-mediated signaling pathway | TAS | 9636140 | |
| GO:0040023 | establishment of nucleus localization | IEA | - | |
| GO:0030198 | extracellular matrix organization | IEA | - | |
| GO:0043542 | endothelial cell migration | IEA | - | |
| GO:0046621 | negative regulation of organ growth | IEA | - | |
| GO:0046777 | protein amino acid autophosphorylation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 9325343 | |
| GO:0005856 | cytoskeleton | IEA | - | |
| GO:0005856 | cytoskeleton | TAS | 10801330 | |
| GO:0005737 | cytoplasm | IDA | 18029348 | |
| GO:0030027 | lamellipodium | IEA | - | |
| GO:0005925 | focal adhesion | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0016324 | apical plasma membrane | IEA | - | |
| GO:0030054 | cell junction | IDA | 18029348 | |
| GO:0030054 | cell junction | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AMPH | AMPH1 | amphiphysin | Affinity Capture-Western | BioGRID | 12558988 |
| APC | BTPS2 | DP2 | DP2.5 | DP3 | GS | adenomatous polyposis coli | - | HPRD | 7744883 |
| ARHGAP26 | FLJ42530 | GRAF | KIAA0621 | OPHN1L | OPHN1L1 | Rho GTPase activating protein 26 | - | HPRD,BioGRID | 8649427 |
| ASAP1 | AMAP1 | CENTB4 | DDEF1 | KIAA1249 | PAG2 | PAP | ZG14P | ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 | - | HPRD | 12058076 |
| ATG12 | APG12 | APG12L | FBR93 | HAPG12 | ATG12 autophagy related 12 homolog (S. cerevisiae) | Two-hybrid | BioGRID | 16189514 |
| BCAR1 | CAS | CAS1 | CASS1 | CRKAS | P130Cas | breast cancer anti-estrogen resistance 1 | - | HPRD | 11514617 |
| BCAR1 | CAS | CAS1 | CASS1 | CRKAS | P130Cas | breast cancer anti-estrogen resistance 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 8649427 |8810278 |11577104 |12135674 |12615911 |
| BCAR1 | CAS | CAS1 | CASS1 | CRKAS | P130Cas | breast cancer anti-estrogen resistance 1 | - | HPRD | 7479864 |11514617 |
| BCAR1 | CAS | CAS1 | CASS1 | CRKAS | P130Cas | breast cancer anti-estrogen resistance 1 | CAS interacts with FAK. This interaction was modeled on a demonstrated interaction between human CAS and mouse FAK. | BIND | 9148935 |
| BIN1 | AMPH2 | AMPHL | DKFZp547F068 | MGC10367 | SH3P9 | bridging integrator 1 | - | HPRD,BioGRID | 12558988 |
| BMX | ETK | PSCTK2 | PSCTK3 | BMX non-receptor tyrosine kinase | - | HPRD,BioGRID | 11331870 |
| CCR5 | CC-CKR-5 | CCCKR5 | CD195 | CKR-5 | CKR5 | CMKBR5 | chemokine (C-C motif) receptor 5 | - | HPRD,BioGRID | 10384144 |
| CD4 | CD4mut | CD4 molecule | - | HPRD | 10384144 |
| CD47 | IAP | MER6 | OA3 | CD47 molecule | - | HPRD | 9169439 |
| CIB1 | CIB | KIP | KIP1 | SIP2-28 | calcium and integrin binding 1 (calmyrin) | - | HPRD,BioGRID | 12881299 |
| CRK | CRKII | v-crk sarcoma virus CT10 oncogene homolog (avian) | Affinity Capture-Western Reconstituted Complex | BioGRID | 10085298 |12615911 |
| CRKL | - | v-crk sarcoma virus CT10 oncogene homolog (avian)-like | Two-hybrid | BioGRID | 15494733 |
| CSK | MGC117393 | c-src tyrosine kinase | - | HPRD,BioGRID | 7529872 |
| CSPG4 | HMW-MAA | MCSP | MCSPG | MEL-CSPG | MSK16 | NG2 | chondroitin sulfate proteoglycan 4 | - | HPRD | 10587647 |
| CXCR4 | CD184 | D2S201E | FB22 | HM89 | HSY3RR | LAP3 | LCR1 | LESTR | NPY3R | NPYR | NPYRL | NPYY3R | WHIM | chemokine (C-X-C motif) receptor 4 | Affinity Capture-Western | BioGRID | 11668182 |
| DCC | CRC18 | CRCR1 | deleted in colorectal carcinoma | Affinity Capture-Western Two-hybrid | BioGRID | 15494733 |
| DCC | CRC18 | CRCR1 | deleted in colorectal carcinoma | DCC interacts with FAK. | BIND | 15494732 |
| DCC | CRC18 | CRCR1 | deleted in colorectal carcinoma | FAK interacts with DCC. | BIND | 15494733 |
| DNM2 | CMTDI1 | CMTDIB | DI-CMTB | DYN2 | DYNII | dynamin 2 | PTK2 (FAK) interacts with DNM2 (DYN2). | BIND | 15895076 |
| DOCK1 | DOCK180 | ced5 | dedicator of cytokinesis 1 | Affinity Capture-Western | BioGRID | 12615911 |
| EFS | CAS3 | CASS3 | EFS1 | EFS2 | HEFS | SIN | embryonal Fyn-associated substrate | - | HPRD | 9750131 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | - | HPRD,BioGRID | 10806474 |
| EPHA2 | ECK | EPH receptor A2 | - | HPRD,BioGRID | 10655584 |
| ERBB2 | CD340 | HER-2 | HER-2/neu | HER2 | NEU | NGL | TKR1 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) | - | HPRD | 11807823 |
| ERBB3 | ErbB-3 | HER3 | LCCS2 | MDA-BF-1 | MGC88033 | c-erbB-3 | c-erbB3 | erbB3-S | p180-ErbB3 | p45-sErbB3 | p85-sErbB3 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian) | - | HPRD | 11807823 |
| EZR | CVIL | CVL | DKFZp762H157 | FLJ26216 | MGC1584 | VIL2 | ezrin | - | HPRD,BioGRID | 11468295 |
| FGR | FLJ43153 | MGC75096 | SRC2 | c-fgr | c-src2 | p55c-fgr | p58c-fgr | Gardner-Rasheed feline sarcoma viral (v-fgr) oncogene homolog | - | HPRD,BioGRID | 12387730 |
| FYN | MGC45350 | SLK | SYN | FYN oncogene related to SRC, FGR, YES | - | HPRD,BioGRID | 11278857 |
| GIT1 | - | G protein-coupled receptor kinase interacting ArfGAP 1 | - | HPRD,BioGRID | 10938112 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | Tyrosine-phosphorylated FAK interacts with Grb2. | BIND | 7597091 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | - | HPRD | 7997267 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | Affinity Capture-Western Reconstituted Complex | BioGRID | 8649427 |10085298 |10806474 |12005431 |12558988 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | Grb2 interacts with FAK. This interaction was modeled on a demonstrated interaction between human Grb2 and mouse FAK. | BIND | 9148935 |
| GRB7 | - | growth factor receptor-bound protein 7 | - | HPRD,BioGRID | 10446223 |
| GSK3B | - | glycogen synthase kinase 3 beta | - | HPRD,BioGRID | 11809746 |
| GSN | DKFZp313L0718 | gelsolin (amyloidosis, Finnish type) | Affinity Capture-Western | BioGRID | 11577104 |
| IGF1R | CD221 | IGFIR | JTK13 | MGC142170 | MGC142172 | MGC18216 | insulin-like growth factor 1 receptor | Affinity Capture-Western | BioGRID | 10082579 |
| IRS1 | HIRS-1 | insulin receptor substrate 1 | - | HPRD,BioGRID | 9822703 |
| ITGA4 | CD49D | IA4 | MGC90518 | integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) | Affinity Capture-Western | BioGRID | 11919182 |
| ITGAV | CD51 | DKFZp686A08142 | MSK8 | VNRA | integrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51) | - | HPRD | 11927607 |
| ITGB1 | CD29 | FNRB | GPIIA | MDF2 | MSK12 | VLA-BETA | VLAB | integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | - | HPRD | 8649427 |
| ITGB2 | CD18 | LAD | LCAMB | LFA-1 | MAC-1 | MF17 | MFI7 | integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) | - | HPRD | 8649427 |
| ITGB3 | CD61 | GP3A | GPIIIa | integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) | FAK interacts with integrin beta 3. | BIND | 11927607 |
| ITGB3 | CD61 | GP3A | GPIIIa | integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) | Affinity Capture-Western Reconstituted Complex | BioGRID | 9169439 |11927607 |
| ITGB3 | CD61 | GP3A | GPIIIa | integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) | - | HPRD | 8649427 |
| ITGB5 | FLJ26658 | integrin, beta 5 | Affinity Capture-Western Reconstituted Complex | BioGRID | 11927607 |
| JAK2 | JTK10 | Janus kinase 2 (a protein tyrosine kinase) | - | HPRD,BioGRID | 9553131 |10925297 |
| KCNMA1 | BKTM | DKFZp686K1437 | KCa1.1 | MGC71881 | MaxiK | SAKCA | SLO | SLO-ALPHA | SLO1 | mSLO1 | potassium large conductance calcium-activated channel, subfamily M, alpha member 1 | - | HPRD | 14584897 |
| LYN | FLJ26625 | JTK8 | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog | Affinity Capture-Western | BioGRID | 9169439 |
| LYN | FLJ26625 | JTK8 | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog | - | HPRD | 11389892 |
| MAPK8IP3 | DKFZp762N1113 | FLJ00027 | JIP3 | JSAP1 | KIAA1066 | SYD2 | mitogen-activated protein kinase 8 interacting protein 3 | Affinity Capture-Western | BioGRID | 12226752 |
| MICAL1 | DKFZp434B1517 | FLJ11937 | FLJ21739 | MICAL | MICAL-1 | NICAL | microtubule associated monoxygenase, calponin and LIM domain containing 1 | - | HPRD | 10808124 |
| NCK1 | MGC12668 | NCK | NCKalpha | NCK adaptor protein 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 8879209 |11950595 |
| NCK2 | GRB4 | NCKbeta | NCK adaptor protein 2 | - | HPRD,BioGRID | 11950595 |
| NEDD9 | CAS-L | CAS2 | CASL | CASS2 | HEF1 | dJ49G10.2 | dJ761I2.1 | neural precursor cell expressed, developmentally down-regulated 9 | - | HPRD,BioGRID | 8668148 |8879209 |10455189 |
| NEO1 | DKFZp547A066 | DKFZp547B146 | HsT17534 | NGN | neogenin homolog 1 (chicken) | FAK interacts with Neogenin. | BIND | 15494733 |
| PCSK1 | BMIQ12 | NEC1 | PC1 | PC3 | SPC3 | proprotein convertase subtilisin/kexin type 1 | Affinity Capture-Western | BioGRID | 11113628 |
| PDGFRB | CD140B | JTK12 | PDGF-R-beta | PDGFR | PDGFR1 | platelet-derived growth factor receptor, beta polypeptide | - | HPRD | 10806474 |
| PIAS1 | DDXBP1 | GBP | GU/RH-II | MGC141878 | MGC141879 | ZMIZ3 | protein inhibitor of activated STAT, 1 | - | HPRD | 14500712 |
| PIK3R1 | GRB1 | p85 | p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | - | HPRD,BioGRID | 7537275 |8649427 |10806474 |
| PKD1 | PBP | polycystic kidney disease 1 (autosomal dominant) | - | HPRD | 11113628 |
| PKD1 | PBP | polycystic kidney disease 1 (autosomal dominant) | polycystin-1 interacts with FAK. | BIND | 11113628 |
| PLCG1 | PLC-II | PLC1 | PLC148 | PLCgamma1 | phospholipase C, gamma 1 | - | HPRD,BioGRID | 8660853|10430888 |
| PPP1CB | MGC3672 | PP-1B | PPP1CD | protein phosphatase 1, catalytic subunit, beta isoform | - | HPRD,BioGRID | 11513739 |
| PTEN | 10q23del | BZS | MGC11227 | MHAM | MMAC1 | PTEN1 | TEP1 | phosphatase and tensin homolog | - | HPRD,BioGRID | 10400703 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | FAK autophosphorylates. | BIND | 11359909 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | - | HPRD,BioGRID | 12005431 |
| PTPN11 | BPTP3 | CFC | MGC14433 | NS1 | PTP-1D | PTP2C | SH-PTP2 | SH-PTP3 | SHP2 | protein tyrosine phosphatase, non-receptor type 11 | - | HPRD,BioGRID | 7589239 |8986614 |10082579 |10655584 |
| PTPN12 | PTP-PEST | PTPG1 | protein tyrosine phosphatase, non-receptor type 12 | - | HPRD | 9497381 |
| PTPRH | FLJ39938 | MGC133058 | MGC133059 | SAP-1 | protein tyrosine phosphatase, receptor type, H | - | HPRD | 11278335 |
| PXN | FLJ16691 | paxillin | FAK interacts with paxillin. | BIND | 11113628 |
| PXN | FLJ16691 | paxillin | - | HPRD,BioGRID | 8922390 |
| RB1CC1 | CC1 | DRAGOU14 | FIP200 | RB1-inducible coiled-coil 1 | - | HPRD,BioGRID | 10769033 |
| RIPK1 | FLJ39204 | RIP | RIP1 | receptor (TNFRSF)-interacting serine-threonine kinase 1 | FAK interacts with RIP. | BIND | 15121855 |
| SELE | CD62E | ELAM | ELAM1 | ESEL | LECAM2 | selectin E | - | HPRD,BioGRID | 8609175 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | - | HPRD,BioGRID | 11980671 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | Shc interacts with FAK. This interaction was modeled on a demonstrated interaction between human Shc and mouse FAK. | BIND | 9148935 |
| SOCS2 | CIS2 | Cish2 | SOCS-2 | SSI-2 | SSI2 | STATI2 | suppressor of cytokine signaling 2 | Two-hybrid | BioGRID | 16189514 |
| SORBS1 | CAP | DKFZp451C066 | DKFZp586P1422 | FLAF2 | FLJ12406 | KIAA1296 | R85FL | SH3D5 | SH3P12 | SORB1 | sorbin and SH3 domain containing 1 | - | HPRD | 9461600 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | c-Src interacts with FAK. This interaction was modeled on a demonstrated interaction between human c-Src and mouse FAK. | BIND | 9148935 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | Affinity Capture-Western Reconstituted Complex | BioGRID | 10085298 |11980671 |12387730 |12558988 |12615911 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | - | HPRD | 7529876 |8054685 |10931187 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | - | HPRD | 7529876 |8054685 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | Src interacts with tyrosine autophosphorylated FAK. This interaction was modeled on a demonstrated interaction between human Src and avian FAK. | BIND | 7515480 |
| STAT1 | DKFZp686B04100 | ISGF-3 | STAT91 | signal transducer and activator of transcription 1, 91kDa | - | HPRD,BioGRID | 11278462 |
| STAT3 | APRF | FLJ20882 | HIES | MGC16063 | signal transducer and activator of transcription 3 (acute-phase response factor) | Affinity Capture-Western | BioGRID | 10925297 |
| SYK | DKFZp313N1010 | FLJ25043 | FLJ37489 | spleen tyrosine kinase | - | HPRD,BioGRID | 9169439 |9342235 |
| TGFB1I1 | ARA55 | HIC-5 | HIC5 | TSC-5 | transforming growth factor beta 1 induced transcript 1 | - | HPRD | 9422762 |9756887 |
| TGFB1I1 | ARA55 | HIC-5 | HIC5 | TSC-5 | transforming growth factor beta 1 induced transcript 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 9422762 |9858471 |11463817 |
| TLN1 | ILWEQ | KIAA1027 | TLN | talin 1 | - | HPRD,BioGRID | 7622520 |
| TRIP6 | MGC10556 | MGC10558 | MGC29959 | MGC3837 | MGC4423 | OIP1 | ZRP-1 | thyroid hormone receptor interactor 6 | - | HPRD,BioGRID | 14688263 |
| TSC1 | KIAA0243 | LAM | MGC86987 | TSC | tuberous sclerosis 1 | Affinity Capture-Western | BioGRID | 17043358 |
| TSC2 | FLJ43106 | LAM | TSC4 | tuberous sclerosis 2 | Affinity Capture-Western Reconstituted Complex | BioGRID | 17043358 |
| YES1 | HsT441 | P61-YES | Yes | c-yes | v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 | Affinity Capture-Western | BioGRID | 12558988 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ERBB SIGNALING PATHWAY | 87 | 71 | All SZGR 2.0 genes in this pathway |
| KEGG CHEMOKINE SIGNALING PATHWAY | 190 | 128 | All SZGR 2.0 genes in this pathway |
| KEGG AXON GUIDANCE | 129 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG VEGF SIGNALING PATHWAY | 76 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG FOCAL ADHESION | 201 | 138 | All SZGR 2.0 genes in this pathway |
| KEGG LEUKOCYTE TRANSENDOTHELIAL MIGRATION | 118 | 78 | All SZGR 2.0 genes in this pathway |
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG SMALL CELL LUNG CANCER | 84 | 67 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AGR PATHWAY | 36 | 31 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AT1R PATHWAY | 36 | 28 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CHEMICAL PATHWAY | 22 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA SPPA PATHWAY | 22 | 18 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CCR3 PATHWAY | 23 | 21 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CELL2CELL PATHWAY | 14 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CXCR4 PATHWAY | 24 | 22 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ECM PATHWAY | 24 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HIVNEF PATHWAY | 58 | 43 | All SZGR 2.0 genes in this pathway |
| BIOCARTA INTEGRIN PATHWAY | 38 | 29 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MCALPAIN PATHWAY | 25 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EDG1 PATHWAY | 27 | 24 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PTEN PATHWAY | 18 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ACH PATHWAY | 16 | 13 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MET PATHWAY | 37 | 30 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TFF PATHWAY | 21 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA UCALPAIN PATHWAY | 18 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA VEGF PATHWAY | 29 | 18 | All SZGR 2.0 genes in this pathway |
| SIG PIP3 SIGNALING IN CARDIAC MYOCTES | 67 | 54 | All SZGR 2.0 genes in this pathway |
| ST GA12 PATHWAY | 23 | 16 | All SZGR 2.0 genes in this pathway |
| ST INTEGRIN SIGNALING PATHWAY | 82 | 62 | All SZGR 2.0 genes in this pathway |
| ST GA13 PATHWAY | 37 | 32 | All SZGR 2.0 genes in this pathway |
| PID FCER1 PATHWAY | 62 | 43 | All SZGR 2.0 genes in this pathway |
| PID LYSOPHOSPHOLIPID PATHWAY | 66 | 53 | All SZGR 2.0 genes in this pathway |
| PID MET PATHWAY | 80 | 60 | All SZGR 2.0 genes in this pathway |
| PID EPHB FWD PATHWAY | 40 | 38 | All SZGR 2.0 genes in this pathway |
| PID NECTIN PATHWAY | 30 | 20 | All SZGR 2.0 genes in this pathway |
| PID RET PATHWAY | 39 | 29 | All SZGR 2.0 genes in this pathway |
| PID ANGIOPOIETIN RECEPTOR PATHWAY | 50 | 41 | All SZGR 2.0 genes in this pathway |
| PID NETRIN PATHWAY | 32 | 27 | All SZGR 2.0 genes in this pathway |
| PID CXCR4 PATHWAY | 102 | 78 | All SZGR 2.0 genes in this pathway |
| PID IGF1 PATHWAY | 30 | 23 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 RECEPTOR PROXIMAL PATHWAY | 35 | 29 | All SZGR 2.0 genes in this pathway |
| PID AVB3 INTEGRIN PATHWAY | 75 | 53 | All SZGR 2.0 genes in this pathway |
| PID SYNDECAN 4 PATHWAY | 32 | 25 | All SZGR 2.0 genes in this pathway |
| PID AR NONGENOMIC PATHWAY | 31 | 27 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 INTERNALIZATION PATHWAY | 41 | 35 | All SZGR 2.0 genes in this pathway |
| PID CASPASE PATHWAY | 52 | 39 | All SZGR 2.0 genes in this pathway |
| PID VEGFR1 2 PATHWAY | 69 | 57 | All SZGR 2.0 genes in this pathway |
| PID EPHA2 FWD PATHWAY | 19 | 16 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN A4B1 PATHWAY | 33 | 24 | All SZGR 2.0 genes in this pathway |
| PID FAK PATHWAY | 59 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | 40 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CELL COMMUNICATION | 120 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRIN CELL SURFACE INTERACTIONS | 79 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | 15 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | 15 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | 27 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | 64 | 49 | All SZGR 2.0 genes in this pathway |
| REACTOME NETRIN1 SIGNALING | 41 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | 13 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | 12 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET AGGREGATION PLUG FORMATION | 36 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOSIS | 148 | 94 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOTIC EXECUTION PHASE | 54 | 37 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL DN | 60 | 39 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| DITTMER PTHLH TARGETS DN | 73 | 51 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 2HR UP | 27 | 19 | All SZGR 2.0 genes in this pathway |
| CHIN BREAST CANCER COPY NUMBER UP | 27 | 18 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B | 392 | 251 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 8Q23 Q24 AMPLICON | 157 | 87 | All SZGR 2.0 genes in this pathway |
| HADDAD T LYMPHOCYTE AND NK PROGENITOR UP | 78 | 56 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D6 | 37 | 25 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| CHIBA RESPONSE TO TSA DN | 23 | 14 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK DN | 318 | 220 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G6 | 153 | 112 | All SZGR 2.0 genes in this pathway |
| YIH RESPONSE TO ARSENITE C1 | 24 | 14 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| LEE METASTASIS AND ALTERNATIVE SPLICING DN | 45 | 31 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D UP | 210 | 124 | All SZGR 2.0 genes in this pathway |
| FUJII YBX1 TARGETS DN | 202 | 132 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| LU TUMOR ANGIOGENESIS UP | 25 | 22 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C D UP | 139 | 95 | All SZGR 2.0 genes in this pathway |
| GU PDEF TARGETS UP | 71 | 49 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER UP | 298 | 174 | All SZGR 2.0 genes in this pathway |
| WENDT COHESIN TARGETS UP | 33 | 19 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE UP | 181 | 106 | All SZGR 2.0 genes in this pathway |
| FONTAINE FOLLICULAR THYROID ADENOMA UP | 75 | 43 | All SZGR 2.0 genes in this pathway |
| FONTAINE THYROID TUMOR UNCERTAIN MALIGNANCY UP | 36 | 23 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 7 | 76 | 46 | All SZGR 2.0 genes in this pathway |
| KARLSSON TGFB1 TARGETS UP | 127 | 78 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS DN | 242 | 146 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 DN | 336 | 211 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP D | 280 | 158 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE MIDDLE | 98 | 56 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-135 | 820 | 826 | m8 | hsa-miR-135a | UAUGGCUUUUUAUUCCUAUGUGA |
| hsa-miR-135b | UAUGGCUUUUCAUUCCUAUGUG | ||||
| miR-138 | 56 | 63 | 1A,m8 | hsa-miR-138brain | AGCUGGUGUUGUGAAUC |
| miR-199 | 844 | 850 | 1A | hsa-miR-199a | CCCAGUGUUCAGACUACCUGUUC |
| hsa-miR-199b | CCCAGUGUUUAGACUAUCUGUUC | ||||
| miR-25/32/92/363/367 | 495 | 501 | 1A | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-378 | 1003 | 1009 | 1A | hsa-miR-378 | CUCCUGACUCCAGGUCCUGUGU |
| miR-410 | 795 | 801 | 1A | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| miR-7 | 875 | 881 | m8 | hsa-miR-7SZ | UGGAAGACUAGUGAUUUUGUUG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.