Gene Page: RPTOR
Summary ?
| GeneID | 57521 |
| Symbol | RPTOR |
| Synonyms | KOG1|Mip1 |
| Description | regulatory associated protein of MTOR, complex 1 |
| Reference | MIM:607130|HGNC:HGNC:30287|Ensembl:ENSG00000141564|HPRD:06184|Vega:OTTHUMG00000177622 |
| Gene type | protein-coding |
| Map location | 17q25.3 |
| Pascal p-value | 3.715E-6 |
| Sherlock p-value | 0.071 |
| Fetal beta | -0.376 |
| DMG | 2 (# studies) |
| eGene | Myers' cis & trans Meta |
| Support | CompositeSet Darnell FMRP targets Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 5 |
| DMG:Montano_2016 | Genome-wide DNA methylation analysis | This dataset includes 172 replicated associations between CpGs with schizophrenia. | 5 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg13549638 | 17 | 78860076 | RPTOR | 6.7E-7 | 0.013 | 0.014 | DMG:Montano_2016 |
| cg27457201 | 17 | 78854232 | RPTOR | 7.92E-6 | 0.008 | 0.038 | DMG:Montano_2016 |
| cg16660971 | 17 | 78860029 | RPTOR | 5.97E-5 | 0.011 | 0.103 | DMG:Montano_2016 |
| cg22091236 | 17 | 78853966 | RPTOR | 1.52E-4 | 0.007 | 0.141 | DMG:Montano_2016 |
| cg22882460 | 17 | 78654648 | RPTOR | 1.43E-8 | 0.022 | 5.52E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17281577 | chr3 | 119051988 | RPTOR | 57521 | 0.17 | trans |
Section II. Transcriptome annotation
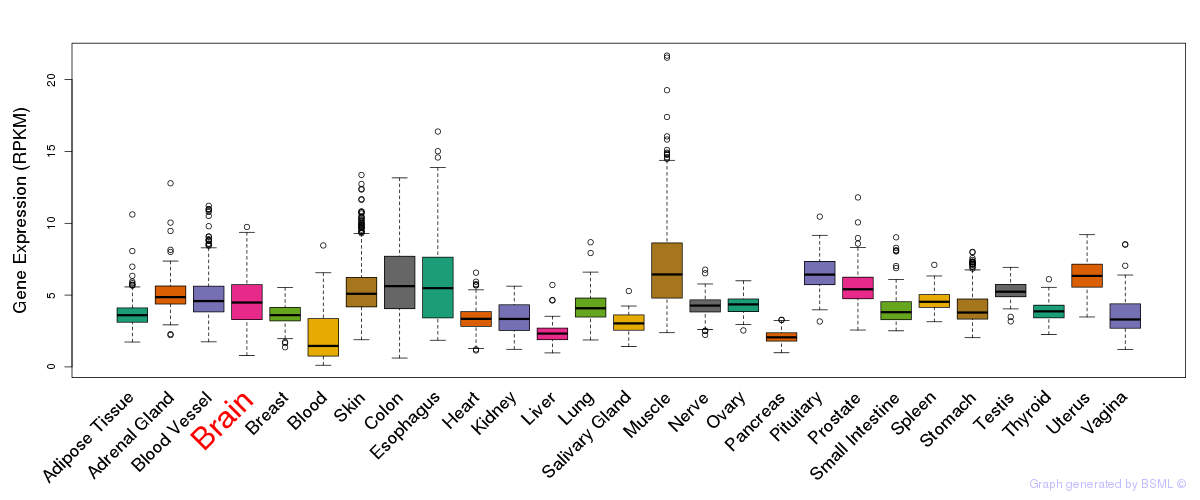
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| EIF3B | EIF3-ETA | EIF3-P110 | EIF3-P116 | EIF3S9 | MGC104664 | MGC131875 | PRT1 | eukaryotic translation initiation factor 3, subunit B | Affinity Capture-Western | BioGRID | 16286006 |
| EIF3C | EIF3S8 | FLJ78287 | MGC189744 | eIF3-p110 | eukaryotic translation initiation factor 3, subunit C | Affinity Capture-Western | BioGRID | 16286006 |
| EIF4EBP1 | 4E-BP1 | 4EBP1 | BP-1 | MGC4316 | PHAS-I | eukaryotic translation initiation factor 4E binding protein 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 12150926 |12604610 |12747827 |12912989 |15767663 |16798736 |16824195 |16837165 |
| FKBP1A | FKBP-12 | FKBP1 | FKBP12 | FKBP12C | PKC12 | PKCI2 | PPIASE | FK506 binding protein 1A, 12kDa | Affinity Capture-Western | BioGRID | 15268862 |15467718 |
| FRAP1 | FLJ44809 | FRAP | FRAP2 | MTOR | RAFT1 | RAPT1 | FK506 binding protein 12-rapamycin associated protein 1 | - | HPRD,BioGRID | 12150925 |
| FRAP1 | FLJ44809 | FRAP | FRAP2 | MTOR | RAFT1 | RAPT1 | FK506 binding protein 12-rapamycin associated protein 1 | mTOR interacts with raptor. | BIND | 15718470 |
| GDI2P | - | GDP dissociation inhibitor 2 pseudogene | Affinity Capture-Western | BioGRID | 16354680 |
| MAPKAP1 | JC310 | MGC2745 | MIP1 | SIN1 | SIN1b | SIN1g | mitogen-activated protein kinase associated protein 1 | Affinity Capture-MS | BioGRID | 16962653 |
| PDK1 | - | pyruvate dehydrogenase kinase, isozyme 1 | Affinity Capture-Western | BioGRID | 12150926 |
| PLD2 | - | phospholipase D2 | Affinity Capture-Western | BioGRID | 16837165 |
| RHEB | MGC111559 | RHEB2 | Ras homolog enriched in brain | Raptor interacts with Rheb. | BIND | 15854902 |
| RHEB | MGC111559 | RHEB2 | Ras homolog enriched in brain | Affinity Capture-Western Reconstituted Complex | BioGRID | 15854902 |
| RICTOR | DKFZp686B11164 | KIAA1999 | MGC39830 | mAVO3 | rapamycin-insensitive companion of mTOR | Affinity Capture-MS | BioGRID | 16962653 |
| RPS6KB1 | PS6K | S6K | S6K1 | STK14A | p70(S6K)-alpha | p70-S6K | p70-alpha | ribosomal protein S6 kinase, 70kDa, polypeptide 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 12150926 |12604610 |15809305 |16837165 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MTOR SIGNALING PATHWAY | 52 | 40 | All SZGR 2.0 genes in this pathway |
| KEGG INSULIN SIGNALING PATHWAY | 137 | 103 | All SZGR 2.0 genes in this pathway |
| PID MET PATHWAY | 80 | 60 | All SZGR 2.0 genes in this pathway |
| PID LKB1 PATHWAY | 47 | 37 | All SZGR 2.0 genes in this pathway |
| PID MTOR 4PATHWAY | 69 | 55 | All SZGR 2.0 genes in this pathway |
| REACTOME INSULIN RECEPTOR SIGNALLING CASCADE | 87 | 64 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | 15 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME ENERGY DEPENDENT REGULATION OF MTOR BY LKB1 AMPK | 18 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME PKB MEDIATED EVENTS | 29 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY INSULIN RECEPTOR | 108 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME MTORC1 MEDIATED SIGNALLING | 11 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K CASCADE | 71 | 51 | All SZGR 2.0 genes in this pathway |
| CORRADETTI MTOR PATHWAY REGULATORS DN | 6 | 5 | All SZGR 2.0 genes in this pathway |
| SILIGAN BOUND BY EWS FLT1 FUSION | 47 | 34 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 17Q21 Q25 AMPLICON | 335 | 181 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| GYORFFY DOXORUBICIN RESISTANCE | 56 | 34 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 | 227 | 149 | All SZGR 2.0 genes in this pathway |