Gene Page: TAOK1
Summary ?
| GeneID | 57551 |
| Symbol | TAOK1 |
| Synonyms | KFC-B|MAP3K16|MARKK|PSK-2|PSK2|TAO1|hKFC-B|hTAOK1 |
| Description | TAO kinase 1 |
| Reference | MIM:610266|HGNC:HGNC:29259|Ensembl:ENSG00000160551|HPRD:15467|Vega:OTTHUMG00000179718 |
| Gene type | protein-coding |
| Map location | 17q11.2 |
| Pascal p-value | 0.53 |
| Sherlock p-value | 5.906E-4 |
| Fetal beta | 0.095 |
| eGene | Myers' cis & trans |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17014160 | chr1 | 206855799 | TAOK1 | 57551 | 0.1 | trans | ||
| rs10915848 | chr1 | 225788919 | TAOK1 | 57551 | 0.16 | trans | ||
| rs7595584 | chr2 | 175652187 | TAOK1 | 57551 | 0.11 | trans | ||
| rs13418573 | chr2 | 205418512 | TAOK1 | 57551 | 0.13 | trans | ||
| rs6804523 | chr3 | 176408204 | TAOK1 | 57551 | 0.16 | trans | ||
| rs6813178 | chr4 | 71575238 | TAOK1 | 57551 | 0.04 | trans | ||
| rs16870339 | chr5 | 2705169 | TAOK1 | 57551 | 0.04 | trans | ||
| rs235427 | chr8 | 133815575 | TAOK1 | 57551 | 0.09 | trans | ||
| rs10115578 | chr9 | 3484372 | TAOK1 | 57551 | 0.17 | trans | ||
| rs4607971 | chr10 | 19407890 | TAOK1 | 57551 | 0.08 | trans | ||
| rs1324669 | chr13 | 107872446 | TAOK1 | 57551 | 0.11 | trans | ||
| rs6015314 | chr20 | 57167224 | TAOK1 | 57551 | 0.14 | trans |
Section II. Transcriptome annotation
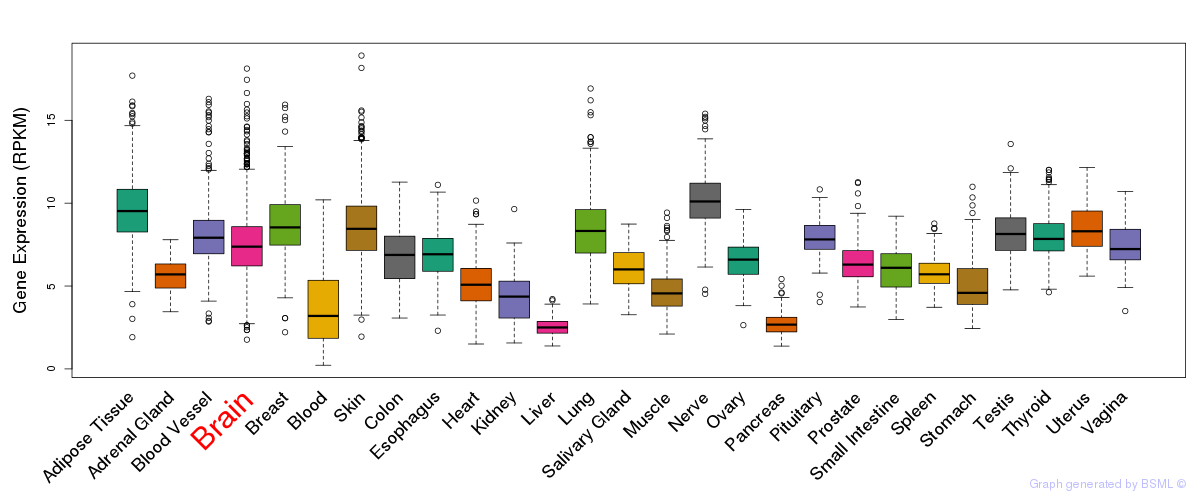
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| PID P38 MKK3 6PATHWAY | 26 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC M M G1 PHASES | 172 | 98 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPLICATION | 192 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC PROMETAPHASE | 87 | 51 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP | 457 | 269 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP DN | 199 | 124 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| SILIGAN TARGETS OF EWS FLI1 FUSION UP | 15 | 12 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 5 6WK DN | 137 | 97 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 6 7WK UP | 197 | 135 | All SZGR 2.0 genes in this pathway |
| FALVELLA SMOKERS WITH LUNG CANCER | 80 | 52 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 17Q11 Q21 AMPLICON | 133 | 78 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| CHANG IMMORTALIZED BY HPV31 UP | 84 | 55 | All SZGR 2.0 genes in this pathway |
| DURCHDEWALD SKIN CARCINOGENESIS DN | 264 | 168 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE UP | 393 | 244 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| FIRESTEIN CTNNB1 PATHWAY AND PROLIFERATION | 9 | 5 | All SZGR 2.0 genes in this pathway |
| FIRESTEIN CTNNB1 PATHWAY | 33 | 23 | All SZGR 2.0 genes in this pathway |
| FIRESTEIN PROLIFERATION | 175 | 125 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS UP | 221 | 135 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| IKEDA MIR30 TARGETS UP | 116 | 87 | All SZGR 2.0 genes in this pathway |
| IKEDA MIR30 TARGETS DN | 27 | 24 | All SZGR 2.0 genes in this pathway |