Gene Page: RAB5B
Summary ?
| GeneID | 5869 |
| Symbol | RAB5B |
| Synonyms | - |
| Description | RAB5B, member RAS oncogene family |
| Reference | MIM:179514|HGNC:HGNC:9784|Ensembl:ENSG00000111540|HPRD:01544|Vega:OTTHUMG00000169682 |
| Gene type | protein-coding |
| Map location | 12q13 |
| Pascal p-value | 0.349 |
| Sherlock p-value | 0.44 |
| Fetal beta | -1.628 |
| DMG | 1 (# studies) |
| Support | INTRACELLULAR TRAFFICKING G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg23594427 | 12 | 56367798 | RAB5B | 1.286E-4 | 0.412 | 0.03 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
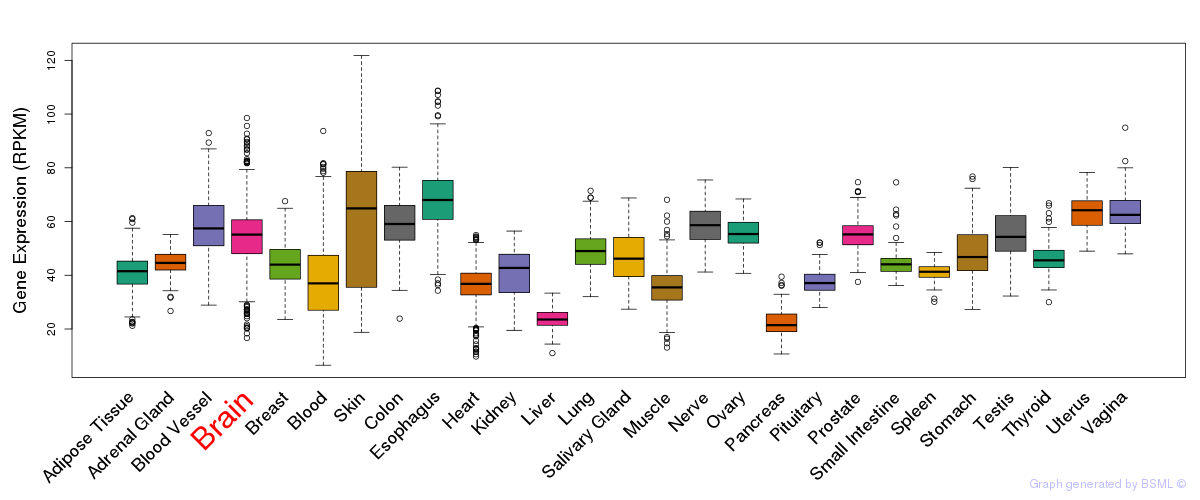
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| XRCC6 | 0.93 | 0.93 |
| CCT7 | 0.92 | 0.90 |
| PRMT5 | 0.92 | 0.88 |
| DRG1 | 0.91 | 0.89 |
| C22orf28 | 0.91 | 0.88 |
| COPZ1 | 0.91 | 0.88 |
| CCT5 | 0.91 | 0.88 |
| MRPL3 | 0.91 | 0.89 |
| ATP5A1 | 0.90 | 0.91 |
| FBXO22 | 0.90 | 0.86 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.72 | -0.69 |
| AF347015.8 | -0.72 | -0.71 |
| AF347015.33 | -0.72 | -0.72 |
| AF347015.26 | -0.71 | -0.72 |
| MT-CYB | -0.71 | -0.71 |
| AF347015.2 | -0.70 | -0.69 |
| AF347015.15 | -0.69 | -0.70 |
| AF347015.31 | -0.69 | -0.68 |
| AF347015.27 | -0.68 | -0.69 |
| AF347015.21 | -0.67 | -0.67 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ENDOCYTOSIS | 183 | 132 | All SZGR 2.0 genes in this pathway |
| KEGG VASOPRESSIN REGULATED WATER REABSORPTION | 44 | 30 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL UP | 450 | 256 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK DN | 196 | 131 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 6 7WK UP | 197 | 135 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 2 | 127 | 92 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| FRIDMAN SENESCENCE UP | 77 | 60 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL FLI1 | 9 | 8 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS UP | 198 | 132 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR DN | 354 | 216 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 12 | 30 | 20 | All SZGR 2.0 genes in this pathway |
| ACOSTA PROLIFERATION INDEPENDENT MYC TARGETS DN | 116 | 74 | All SZGR 2.0 genes in this pathway |
| LIU IL13 PRIMING MODEL | 15 | 11 | All SZGR 2.0 genes in this pathway |
| ALFANO MYC TARGETS | 239 | 156 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL DN | 428 | 246 | All SZGR 2.0 genes in this pathway |