Gene Page: RYR3
Summary ?
| GeneID | 6263 |
| Symbol | RYR3 |
| Synonyms | RYR-3 |
| Description | ryanodine receptor 3 |
| Reference | MIM:180903|HGNC:HGNC:10485|Ensembl:ENSG00000198838|HPRD:01620|Vega:OTTHUMG00000172253 |
| Gene type | protein-coding |
| Map location | 15q13.3 |
| Pascal p-value | 0.015 |
| Sherlock p-value | 0.69 |
| TADA p-value | 0.007 |
| Fetal beta | -0.426 |
| Support | INTRACELLULAR SIGNAL TRANSDUCTION CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| RYR3 | chr15 | 33999249 | G | A | NM_001036 NM_001243996 | p.2205V>M p.2205V>M | missense missense | Schizophrenia | DNM:Fromer_2014 | ||
| RYR3 | chr15 | 34151822 | T | C | NM_001036 NM_001243996 | p.4730I>T p.4725I>T | missense missense | Schizophrenia | DNM:Fromer_2014 |
Section II. Transcriptome annotation
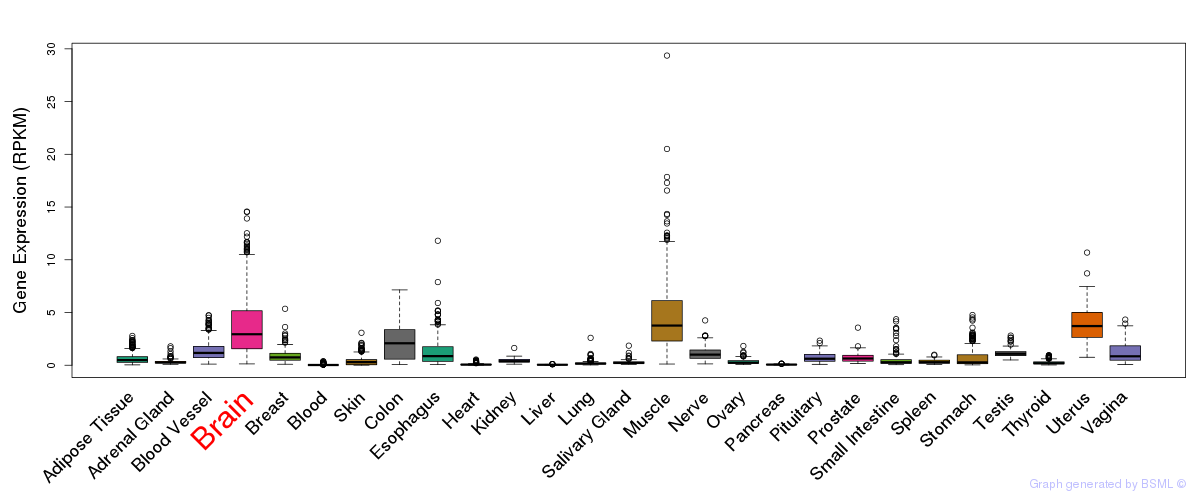
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DDR1 | 0.90 | 0.90 |
| HEATR2 | 0.87 | 0.87 |
| LIX1L | 0.87 | 0.91 |
| FHL1 | 0.86 | 0.86 |
| GOLM1 | 0.86 | 0.86 |
| DAG1 | 0.86 | 0.86 |
| AC130686.2 | 0.85 | 0.84 |
| SEPN1 | 0.85 | 0.89 |
| FYN | 0.85 | 0.88 |
| FAM57A | 0.85 | 0.83 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.65 | -0.71 |
| AF347015.31 | -0.63 | -0.75 |
| MT-CO2 | -0.63 | -0.75 |
| AF347015.27 | -0.61 | -0.73 |
| CA4 | -0.61 | -0.68 |
| AF347015.33 | -0.60 | -0.71 |
| MYL3 | -0.60 | -0.72 |
| FXYD1 | -0.59 | -0.70 |
| AF347015.21 | -0.59 | -0.78 |
| COX4I2 | -0.59 | -0.69 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CALCIUM SIGNALING PATHWAY | 178 | 134 | All SZGR 2.0 genes in this pathway |
| KEGG ALZHEIMERS DISEASE | 169 | 110 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS UP | 332 | 228 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST LOBULAR CARCINOMA VS LOBULAR NORMAL UP | 94 | 59 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL UP | 276 | 187 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP | 390 | 236 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| ROSS AML OF FAB M7 TYPE | 68 | 44 | All SZGR 2.0 genes in this pathway |
| YAGI AML FAB MARKERS | 191 | 131 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D UP | 210 | 124 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |