Gene Page: BHMT
Summary ?
| GeneID | 635 |
| Symbol | BHMT |
| Synonyms | BHMT1|HEL-S-61p |
| Description | betaine--homocysteine S-methyltransferase |
| Reference | MIM:602888|HGNC:HGNC:1047|Ensembl:ENSG00000145692|HPRD:04200|Vega:OTTHUMG00000108157 |
| Gene type | protein-coding |
| Map location | 5q14.1 |
| Pascal p-value | 0.033 |
| eGene | Caudate basal ganglia Cortex Hippocampus Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
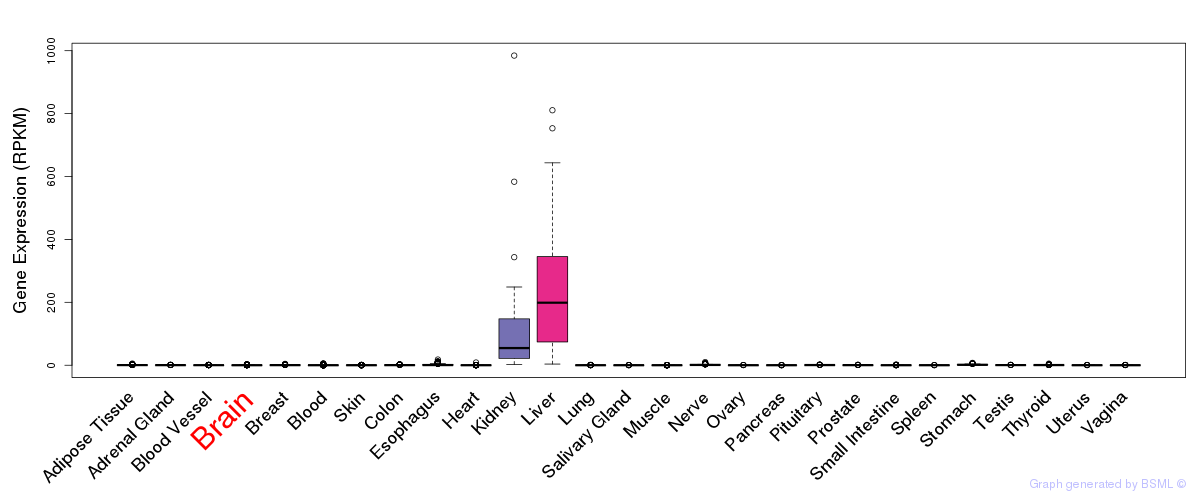
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AC021087.1 | 0.53 | 0.28 |
| GSX1 | 0.51 | 0.19 |
| C1QL4 | 0.51 | 0.12 |
| PRSS36 | 0.51 | 0.25 |
| PROK2 | 0.50 | 0.25 |
| PRG2 | 0.50 | 0.14 |
| SLC5A10 | 0.49 | 0.20 |
| KISS1R | 0.48 | 0.15 |
| C3orf45 | 0.47 | 0.19 |
| MTL5 | 0.46 | 0.10 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| COX4I2 | -0.22 | -0.29 |
| C5orf53 | -0.22 | -0.26 |
| MT-CO2 | -0.21 | -0.27 |
| AF347015.31 | -0.21 | -0.28 |
| AF347015.33 | -0.20 | -0.26 |
| MOBP | -0.20 | -0.24 |
| AF347015.27 | -0.20 | -0.27 |
| MT-CYB | -0.20 | -0.27 |
| MBP | -0.19 | -0.22 |
| GBP2 | -0.18 | -0.22 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG GLYCINE SERINE AND THREONINE METABOLISM | 31 | 26 | All SZGR 2.0 genes in this pathway |
| KEGG CYSTEINE AND METHIONINE METABOLISM | 34 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME SULFUR AMINO ACID METABOLISM | 24 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | 200 | 136 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 48HR UP | 128 | 95 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 96HR UP | 117 | 84 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM4 | 261 | 153 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER CIPROFIBRATE DN | 66 | 43 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC TGFA DN | 65 | 44 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC E2F1 DN | 64 | 38 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER DENA DN | 74 | 45 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC DN | 63 | 40 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER E2F1 DN | 64 | 42 | All SZGR 2.0 genes in this pathway |
| HSIAO LIVER SPECIFIC GENES | 244 | 154 | All SZGR 2.0 genes in this pathway |
| KUNINGER IGF1 VS PDGFB TARGETS DN | 46 | 28 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS PROLIFERATION DN | 179 | 97 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS UNANNOTATED UP | 85 | 50 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S3 | 266 | 180 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT DN | 222 | 141 | All SZGR 2.0 genes in this pathway |
| SUMI HNF4A TARGETS | 34 | 14 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |