Gene Page: BID
Summary ?
| GeneID | 637 |
| Symbol | BID |
| Synonyms | FP497 |
| Description | BH3 interacting domain death agonist |
| Reference | MIM:601997|HGNC:HGNC:1050|Ensembl:ENSG00000015475|HPRD:03590|Vega:OTTHUMG00000150087 |
| Gene type | protein-coding |
| Map location | 22q11.1 |
| Pascal p-value | 0.507 |
| Sherlock p-value | 0.189 |
| Fetal beta | 0.602 |
| eGene | Caudate basal ganglia Cerebellum Cortex Frontal Cortex BA9 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.031 | |
| Expression | Meta-analysis of gene expression | P value: 1.541 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
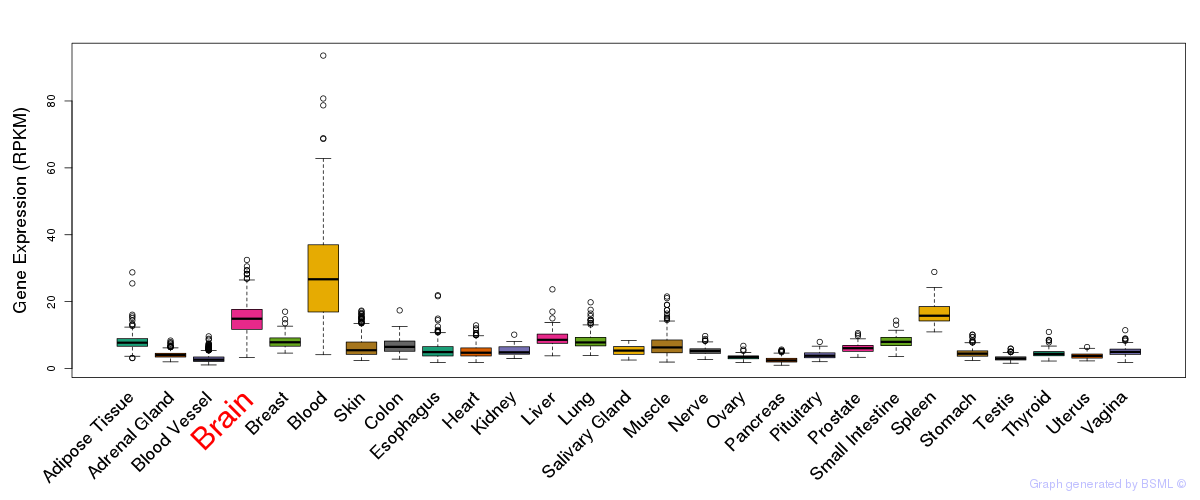
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005123 | death receptor binding | TAS | 8918887 | |
| GO:0005515 | protein binding | ISS | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0051402 | neuron apoptosis | TAS | neuron, axon, dendrite (GO term level: 7) | 16167175 |
| GO:0001836 | release of cytochrome c from mitochondria | IDA | 17052454 | |
| GO:0008625 | induction of apoptosis via death domain receptors | TAS | 9727491 | |
| GO:0008633 | activation of pro-apoptotic gene products | EXP | 15231831 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 10629050 |11099414 |11114298 |12804595 | |
| GO:0005829 | cytosol | TAS | 8918887 | |
| GO:0005624 | membrane fraction | TAS | 8918887 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005739 | mitochondrion | TAS | 9727491 | |
| GO:0005741 | mitochondrial outer membrane | EXP | 9727492 |10950869 |12624108 | |
| GO:0016020 | membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ARHGDIA | GDIA1 | MGC117248 | RHOGDI | RHOGDI-1 | Rho GDP dissociation inhibitor (GDI) alpha | Co-purification | BioGRID | 15659383 |
| BAK1 | BAK | BAK-LIKE | BCL2L7 | CDN1 | MGC117255 | MGC3887 | BCL2-antagonist/killer 1 | - | HPRD | 10950869 |
| BAX | BCL2L4 | BCL2-associated X protein | Bax interacts with Bid. | BIND | 15574335 |
| BCL2 | Bcl-2 | B-cell CLL/lymphoma 2 | Bid interacts with Bcl-2. | BIND | 15694340 |
| BCL2 | Bcl-2 | B-cell CLL/lymphoma 2 | Protein-peptide | BioGRID | 15520201 |15694340 |
| BCL2 | Bcl-2 | B-cell CLL/lymphoma 2 | Bid interacts with Bcl-2. | BIND | 15574335 |
| BCL2 | Bcl-2 | B-cell CLL/lymphoma 2 | - | HPRD | 9727491 |
| BCL2A1 | ACC-1 | ACC-2 | BCL2L5 | BFL1 | GRS | HBPA1 | BCL2-related protein A1 | Bid interacts with A1. This interaction was modelled on a demonstrated interaction between human Bid and mouse A1. | BIND | 15694340 |
| BCL2A1 | ACC-1 | ACC-2 | BCL2L5 | BFL1 | GRS | HBPA1 | BCL2-related protein A1 | - | HPRD,BioGRID | 11929871 |
| BCL2L1 | BCL-XL/S | BCL2L | BCLX | Bcl-X | DKFZp781P2092 | bcl-xL | bcl-xS | BCL2-like 1 | Bid interacts with Bcl-XL. | BIND | 15694340 |
| BCL2L1 | BCL-XL/S | BCL2L | BCLX | Bcl-X | DKFZp781P2092 | bcl-xL | bcl-xS | BCL2-like 1 | Protein-peptide | BioGRID | 15694340 |
| BCL2L1 | BCL-XL/S | BCL2L | BCLX | Bcl-X | DKFZp781P2092 | bcl-xL | bcl-xS | BCL2-like 1 | BCL-XL interacts with the alpha-helical region of the BID-EL BH3 death domain. | BIND | 15353804 |
| BCL2L1 | BCL-XL/S | BCL2L | BCLX | Bcl-X | DKFZp781P2092 | bcl-xL | bcl-xS | BCL2-like 1 | BCL-XL interacts with the alpha-helical region of the BID BH3 death domain. | BIND | 15353804 |
| BCL2L1 | BCL-XL/S | BCL2L | BCLX | Bcl-X | DKFZp781P2092 | bcl-xL | bcl-xS | BCL2-like 1 | Bcl-XL interacts with Bid. This interaction was modelled on a demonstrated interaction between Bcl-XL from an unspecified species and human Bid. | BIND | 15721256 |
| BCL2L1 | BCL-XL/S | BCL2L | BCLX | Bcl-X | DKFZp781P2092 | bcl-xL | bcl-xS | BCL2-like 1 | Bid interacts with Bcl-xL. | BIND | 15574335 |
| BCL2L2 | BCL-W | BCLW | KIAA0271 | BCL2-like 2 | Protein-peptide | BioGRID | 15694340 |
| BCL2L2 | BCL-W | BCLW | KIAA0271 | BCL2-like 2 | Bid interacts with Bcl-w. | BIND | 15694340 |
| BID | FP497 | MGC15319 | MGC42355 | BH3 interacting domain death agonist | - | HPRD,BioGRID | 11805084 |
| BNC1 | BNC | BSN1 | HsT19447 | basonuclin 1 | Bid interacts with Mcl-1. This interaction was modelled on a demonstrated interaction between human Bid and mouse Mcl-1. | BIND | 15694340 |
| CAPN1 | CANP | CANP1 | CANPL1 | muCANP | muCL | calpain 1, (mu/I) large subunit | - | HPRD | 12000759 |
| CAPN2 | CANP2 | CANPL2 | CANPml | FLJ39928 | mCANP | calpain 2, (m/II) large subunit | Biochemical Activity | BioGRID | 12000759 |
| CASP10 | ALPS2 | FLICE2 | MCH4 | caspase 10, apoptosis-related cysteine peptidase | Co-purification | BioGRID | 15659383 |
| CASP2 | CASP-2 | ICH-1L | ICH-1L/1S | ICH1 | NEDD2 | caspase 2, apoptosis-related cysteine peptidase | - | HPRD,BioGRID | 11832478 |
| CASP3 | CPP32 | CPP32B | SCA-1 | caspase 3, apoptosis-related cysteine peptidase | Biochemical Activity | BioGRID | 11399776 |
| CASP8 | ALPS2B | CAP4 | FLICE | FLJ17672 | MACH | MCH5 | MGC78473 | caspase 8, apoptosis-related cysteine peptidase | Biochemical Activity Co-purification | BioGRID | 11832478 |15659383 |
| EZR | CVIL | CVL | DKFZp762H157 | FLJ26216 | MGC1584 | VIL2 | ezrin | Co-purification | BioGRID | 15659383 |
| FADD | GIG3 | MGC8528 | MORT1 | Fas (TNFRSF6)-associated via death domain | Co-purification | BioGRID | 15659383 |
| FAS | ALPS1A | APO-1 | APT1 | CD95 | FAS1 | FASTM | TNFRSF6 | Fas (TNF receptor superfamily, member 6) | Co-purification | BioGRID | 15659383 |
| FASLG | APT1LG1 | CD178 | CD95L | FASL | TNFSF6 | Fas ligand (TNF superfamily, member 6) | Co-purification | BioGRID | 15659383 |
| MAPK8 | JNK | JNK1 | JNK1A2 | JNK21B1/2 | PRKM8 | SAPK1 | mitogen-activated protein kinase 8 | Co-purification | BioGRID | 15659383 |
| MCL1 | BCL2L3 | EAT | MCL1L | MCL1S | MGC104264 | MGC1839 | TM | myeloid cell leukemia sequence 1 (BCL2-related) | Affinity Capture-Western Protein-peptide Reconstituted Complex | BioGRID | 15637055 |15694340 |
| MSN | - | moesin | Co-purification | BioGRID | 15659383 |
| PACS2 | FLJ25488 | KIAA0602 | PACS1L | phosphofurin acidic cluster sorting protein 2 | PACS2 interacts with BID. | BIND | 15692567 |
| RHOA | ARH12 | ARHA | RHO12 | RHOH12 | ras homolog gene family, member A | Co-purification | BioGRID | 15659383 |
| TNFRSF10B | CD262 | DR5 | KILLER | KILLER/DR5 | TRAIL-R2 | TRAILR2 | TRICK2 | TRICK2A | TRICK2B | TRICKB | ZTNFR9 | tumor necrosis factor receptor superfamily, member 10b | Co-purification | BioGRID | 15659383 |
| TNFRSF1A | CD120a | FPF | MGC19588 | TBP1 | TNF-R | TNF-R-I | TNF-R55 | TNFAR | TNFR1 | TNFR55 | TNFR60 | p55 | p55-R | p60 | tumor necrosis factor receptor superfamily, member 1A | Co-purification | BioGRID | 15659383 |
| YWHAB | GW128 | HS1 | KCIP-1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | - | HPRD | 12871587 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG P53 SIGNALING PATHWAY | 69 | 45 | All SZGR 2.0 genes in this pathway |
| KEGG APOPTOSIS | 88 | 62 | All SZGR 2.0 genes in this pathway |
| KEGG NATURAL KILLER CELL MEDIATED CYTOTOXICITY | 137 | 92 | All SZGR 2.0 genes in this pathway |
| KEGG ALZHEIMERS DISEASE | 169 | 110 | All SZGR 2.0 genes in this pathway |
| KEGG AMYOTROPHIC LATERAL SCLEROSIS ALS | 53 | 43 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG VIRAL MYOCARDITIS | 73 | 58 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CHEMICAL PATHWAY | 22 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HIVNEF PATHWAY | 58 | 43 | All SZGR 2.0 genes in this pathway |
| BIOCARTA DEATH PATHWAY | 33 | 24 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MITOCHONDRIA PATHWAY | 21 | 15 | All SZGR 2.0 genes in this pathway |
| SA PROGRAMMED CELL DEATH | 12 | 9 | All SZGR 2.0 genes in this pathway |
| PID ATM PATHWAY | 34 | 25 | All SZGR 2.0 genes in this pathway |
| PID FAS PATHWAY | 38 | 29 | All SZGR 2.0 genes in this pathway |
| PID CERAMIDE PATHWAY | 48 | 37 | All SZGR 2.0 genes in this pathway |
| PID P53 DOWNSTREAM PATHWAY | 137 | 94 | All SZGR 2.0 genes in this pathway |
| PID HIV NEF PATHWAY | 35 | 26 | All SZGR 2.0 genes in this pathway |
| PID CASPASE PATHWAY | 52 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOSIS | 148 | 94 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF BH3 ONLY PROTEINS | 17 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | 30 | 21 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP | 579 | 346 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY UP | 430 | 232 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 UP | 380 | 236 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 UP | 418 | 263 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 48HR UP | 128 | 95 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 96HR UP | 117 | 84 | All SZGR 2.0 genes in this pathway |
| LAU APOPTOSIS CDKN2A UP | 55 | 40 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 DN | 378 | 231 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF UP | 215 | 137 | All SZGR 2.0 genes in this pathway |
| DARWICHE SKIN TUMOR PROMOTER DN | 185 | 115 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK LOW DN | 165 | 107 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK HIGH UP | 147 | 101 | All SZGR 2.0 genes in this pathway |
| DARWICHE SQUAMOUS CELL CARCINOMA UP | 146 | 104 | All SZGR 2.0 genes in this pathway |
| RAMJAUN APOPTOSIS BY TGFB1 VIA MAPK1 DN | 8 | 6 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| GALLUZZI PERMEABILIZE MITOCHONDRIA | 43 | 31 | All SZGR 2.0 genes in this pathway |
| ROSTY CERVICAL CANCER PROLIFERATION CLUSTER | 140 | 73 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO IKK INHIBITOR AND TNF UP | 223 | 140 | All SZGR 2.0 genes in this pathway |
| STANELLE E2F1 TARGETS | 29 | 20 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| HESS TARGETS OF HOXA9 AND MEIS1 UP | 65 | 44 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| TRAYNOR RETT SYNDROM UP | 45 | 33 | All SZGR 2.0 genes in this pathway |
| GERHOLD ADIPOGENESIS DN | 64 | 44 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS INTERMEDIATE PROGENITOR | 149 | 84 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 24HR UP | 148 | 96 | All SZGR 2.0 genes in this pathway |
| BILD MYC ONCOGENIC SIGNATURE | 206 | 117 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| HOWLIN CITED1 TARGETS 2 DN | 17 | 11 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS REQUIRE MYC | 210 | 123 | All SZGR 2.0 genes in this pathway |
| LEE METASTASIS AND ALTERNATIVE SPLICING UP | 74 | 51 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC | 420 | 269 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| VANASSE BCL2 TARGETS DN | 74 | 50 | All SZGR 2.0 genes in this pathway |
| COLINA TARGETS OF 4EBP1 AND 4EBP2 | 356 | 214 | All SZGR 2.0 genes in this pathway |
| MOOTHA GLUCONEOGENESIS | 32 | 23 | All SZGR 2.0 genes in this pathway |
| MOOTHA GLYCOLYSIS | 21 | 13 | All SZGR 2.0 genes in this pathway |
| LI DCP2 BOUND MRNA | 89 | 57 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP D | 280 | 158 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS UP | 279 | 155 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE VIA P38 PARTIAL | 160 | 106 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38 | 337 | 236 | All SZGR 2.0 genes in this pathway |
| RATTENBACHER BOUND BY CELF1 | 467 | 251 | All SZGR 2.0 genes in this pathway |