Gene Page: BLK
Summary ?
| GeneID | 640 |
| Symbol | BLK |
| Synonyms | MODY11 |
| Description | BLK proto-oncogene, Src family tyrosine kinase |
| Reference | MIM:191305|HGNC:HGNC:1057|Ensembl:ENSG00000136573|HPRD:01866|Vega:OTTHUMG00000090729 |
| Gene type | protein-coding |
| Map location | 8p23-p22 |
| Pascal p-value | 1.997E-4 |
| Fetal beta | 0.114 |
| DMG | 2 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg22826986 | 8 | 11351507 | BLK | 3.256E-4 | -0.735 | 0.04 | DMG:Wockner_2014 |
| cg03860768 | 8 | 11351571 | BLK | 2.71E-6 | -7.301 | DMG:vanEijk_2014 |
Section II. Transcriptome annotation
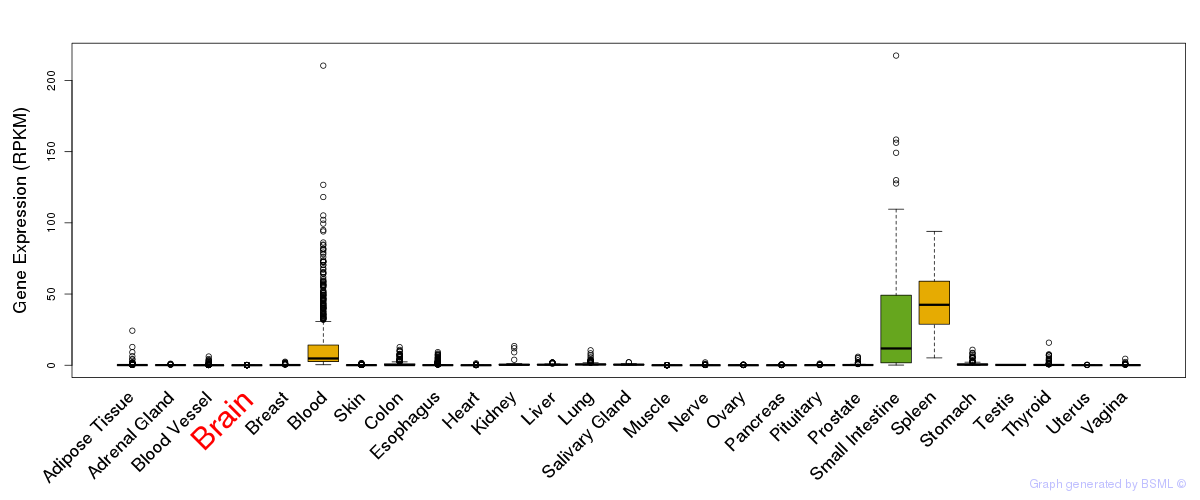
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BCAS2 | DAM1 | breast carcinoma amplified sequence 2 | - | HPRD | 12406557 |
| BCL2 | Bcl-2 | B-cell CLL/lymphoma 2 | - | HPRD,BioGRID | 9525867 |
| BCL2L1 | BCL-XL/S | BCL2L | BCLX | Bcl-X | DKFZp781P2092 | bcl-xL | bcl-xS | BCL2-like 1 | Reconstituted Complex | BioGRID | 9525867 |
| BCR | ALL | BCR-ABL1 | BCR1 | CML | D22S11 | D22S662 | FLJ16453 | PHL | breakpoint cluster region | - | HPRD | 8756631 |
| CBL | C-CBL | CBL2 | RNF55 | Cas-Br-M (murine) ecotropic retroviral transforming sequence | - | HPRD | 8083187 |
| CD79A | IGA | MB-1 | CD79a molecule, immunoglobulin-associated alpha | - | HPRD,BioGRID | 7592958 |
| CD79B | B29 | IGB | CD79b molecule, immunoglobulin-associated beta | - | HPRD,BioGRID | 7592958 |
| CTR9 | KIAA0155 | SH2BP1 | TSBP | p150 | p150TSP | Ctr9, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) | - | HPRD | 8636124 |
| FCGR2A | CD32 | CD32A | CDw32 | FCG2 | FCGR2 | FCGR2A1 | FcGR | IGFR2 | MGC23887 | MGC30032 | Fc fragment of IgG, low affinity IIa, receptor (CD32) | - | HPRD | 8756631 |
| PLCG2 | - | phospholipase C, gamma 2 (phosphatidylinositol-specific) | - | HPRD,BioGRID | 8395016 |
| UBE3A | ANCR | AS | E6-AP | EPVE6AP | FLJ26981 | HPVE6A | ubiquitin protein ligase E3A | - | HPRD,BioGRID | 10449731 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID GLYPICAN 1PATHWAY | 27 | 20 | All SZGR 2.0 genes in this pathway |
| PID PTP1B PATHWAY | 52 | 40 | All SZGR 2.0 genes in this pathway |
| PID P38 ALPHA BETA PATHWAY | 31 | 25 | All SZGR 2.0 genes in this pathway |
| PID TXA2PATHWAY | 57 | 43 | All SZGR 2.0 genes in this pathway |
| PID CXCR4 PATHWAY | 102 | 78 | All SZGR 2.0 genes in this pathway |
| PID PI3KCI PATHWAY | 49 | 40 | All SZGR 2.0 genes in this pathway |
| PID AMB2 NEUTROPHILS PATHWAY | 41 | 32 | All SZGR 2.0 genes in this pathway |
| PID EPHA FWDPATHWAY | 34 | 29 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PID EPHRINB REV PATHWAY | 30 | 25 | All SZGR 2.0 genes in this pathway |
| PID ALPHA SYNUCLEIN PATHWAY | 33 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | 29 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | 126 | 90 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 UP | 201 | 125 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL UP | 276 | 187 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| MORI PRE BI LYMPHOCYTE DN | 77 | 49 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION UP | 38 | 25 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS B LYMPHOCYTE DN | 38 | 25 | All SZGR 2.0 genes in this pathway |
| HOFMANN CELL LYMPHOMA UP | 50 | 35 | All SZGR 2.0 genes in this pathway |
| YU MYC TARGETS DN | 55 | 38 | All SZGR 2.0 genes in this pathway |
| HADDAD B LYMPHOCYTE PROGENITOR | 293 | 193 | All SZGR 2.0 genes in this pathway |
| GREENBAUM E2A TARGETS DN | 21 | 11 | All SZGR 2.0 genes in this pathway |
| KUMAR TARGETS OF MLL AF9 FUSION | 405 | 264 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 10HR DN | 56 | 37 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 24HR DN | 148 | 102 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN | 298 | 200 | All SZGR 2.0 genes in this pathway |
| EHRLICH ICF SYNDROM DN | 15 | 13 | All SZGR 2.0 genes in this pathway |
| MCCLUNG COCAIN REWARD 4WK | 75 | 47 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 48HR DN | 161 | 105 | All SZGR 2.0 genes in this pathway |
| HELLER SILENCED BY METHYLATION UP | 282 | 183 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA UP | 259 | 185 | All SZGR 2.0 genes in this pathway |
| ZHAN EARLY DIFFERENTIATION GENES DN | 42 | 29 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 1HR DN | 222 | 147 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP D | 280 | 158 | All SZGR 2.0 genes in this pathway |