Gene Page: SHB
Summary ?
| GeneID | 6461 |
| Symbol | SHB |
| Synonyms | bA3J10.2 |
| Description | Src homology 2 domain containing adaptor protein B |
| Reference | MIM:600314|HGNC:HGNC:10838|HPRD:02635| |
| Gene type | protein-coding |
| Map location | 9p13.2 |
| Pascal p-value | 0.004 |
| Fetal beta | 2.207 |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
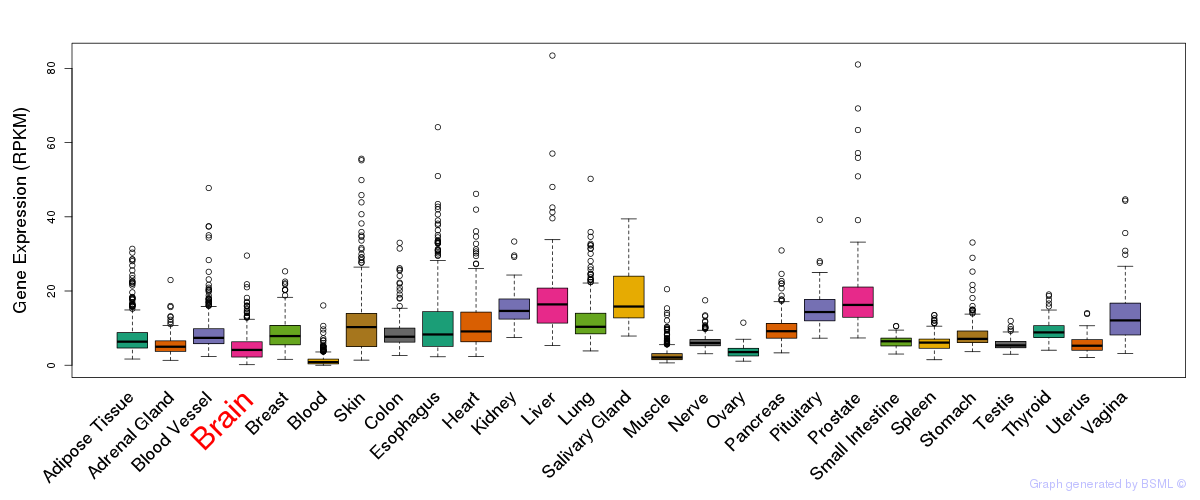
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NLGN2 | 0.93 | 0.94 |
| APC2 | 0.92 | 0.94 |
| PRR12 | 0.92 | 0.93 |
| CHERP | 0.92 | 0.93 |
| ARHGEF18 | 0.91 | 0.93 |
| AC084125.1 | 0.91 | 0.92 |
| CLIP2 | 0.91 | 0.93 |
| MNT | 0.91 | 0.92 |
| BICD2 | 0.91 | 0.92 |
| FOXK2 | 0.91 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.72 | -0.79 |
| MT-CO2 | -0.70 | -0.79 |
| S100B | -0.69 | -0.75 |
| AF347015.27 | -0.69 | -0.75 |
| COPZ2 | -0.68 | -0.73 |
| AF347015.33 | -0.68 | -0.73 |
| FXYD1 | -0.68 | -0.73 |
| AF347015.21 | -0.68 | -0.83 |
| HIGD1B | -0.67 | -0.76 |
| MT-CYB | -0.67 | -0.74 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CRK | CRKII | v-crk sarcoma virus CT10 oncogene homolog (avian) | - | HPRD,BioGRID | 10964504 |
| EPS8 | - | epidermal growth factor receptor pathway substrate 8 | Shb interacts with Eps8. This interaction was modelled on a demonstrated interaction between human Shb and murine Eps8 | BIND | 7537362 |
| EPS8 | - | epidermal growth factor receptor pathway substrate 8 | - | HPRD,BioGRID | 7537362 |
| FGFR1 | BFGFR | CD331 | CEK | FGFBR | FLG | FLJ99988 | FLT2 | HBGFR | KAL2 | N-SAM | fibroblast growth factor receptor 1 | - | HPRD,BioGRID | 7537362 |
| FGFR1 | BFGFR | CD331 | CEK | FGFBR | FLG | FLJ99988 | FLT2 | HBGFR | KAL2 | N-SAM | fibroblast growth factor receptor 1 | Shb interacts with FGFR1 | BIND | 7537362 |
| GRAP2 | GADS | GRAP-2 | GRB2L | GRBLG | GRID | GRPL | GrbX | Grf40 | Mona | P38 | GRB2-related adaptor protein 2 | - | HPRD | 12084069 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | - | HPRD,BioGRID | 9484780 |10488157 |
| IL2RB | CD122 | P70-75 | interleukin 2 receptor, beta | - | HPRD,BioGRID | 12200137 |
| IL2RG | CD132 | IMD4 | SCIDX | SCIDX1 | interleukin 2 receptor, gamma (severe combined immunodeficiency) | - | HPRD,BioGRID | 12200137 |
| JAK1 | JAK1A | JAK1B | JTK3 | Janus kinase 1 (a protein tyrosine kinase) | - | HPRD,BioGRID | 12200137 |
| JAK3 | JAK-3 | JAK3_HUMAN | JAKL | L-JAK | LJAK | Janus kinase 3 (a protein tyrosine kinase, leukocyte) | - | HPRD,BioGRID | 12200137 |
| KDR | CD309 | FLK1 | VEGFR | VEGFR2 | kinase insert domain receptor (a type III receptor tyrosine kinase) | - | HPRD,BioGRID | 15026417 |
| LAT | LAT1 | pp36 | linker for activation of T cells | - | HPRD,BioGRID | 12084069 |
| LCP2 | SLP-76 | SLP76 | lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) | - | HPRD,BioGRID | 12084069 |
| PDGFRA | CD140A | MGC74795 | PDGFR2 | Rhe-PDGFRA | platelet-derived growth factor receptor, alpha polypeptide | - | HPRD | 10837138 |
| PDGFRB | CD140B | JTK12 | PDGF-R-beta | PDGFR | PDGFR1 | platelet-derived growth factor receptor, beta polypeptide | Shb interacts with the PDGF beta-receptor. | BIND | 7537362 |
| PIK3R1 | GRB1 | p85 | p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | Shb interacts with p85-alpha PI3 kinase. This interaction was modelled on a demonstrated interaction between human Shb and bovine p85-alpha PI3 kinase. | BIND | 7537362 |
| PIK3R1 | GRB1 | p85 | p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | - | HPRD,BioGRID | 7537362 |
| PLCG1 | PLC-II | PLC1 | PLC148 | PLCgamma1 | phospholipase C, gamma 1 | Affinity Capture-Western | BioGRID | 10488157 |
| PLCG1 | PLC-II | PLC1 | PLC148 | PLCgamma1 | phospholipase C, gamma 1 | - | HPRD | 12084069 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | Shb interacts with Src. This interaction was modelled on a demonstrated interaction between human Shb and chicken Src. | BIND | 7537362 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | - | HPRD,BioGRID | 7537362 |
| VAV1 | VAV | vav 1 guanine nucleotide exchange factor | - | HPRD,BioGRID | 12084069 |
| ZAP70 | FLJ17670 | FLJ17679 | SRK | STD | TZK | ZAP-70 | zeta-chain (TCR) associated protein kinase 70kDa | - | HPRD,BioGRID | 12084069 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID PDGFRA PATHWAY | 22 | 18 | All SZGR 2.0 genes in this pathway |
| PID VEGFR1 2 PATHWAY | 69 | 57 | All SZGR 2.0 genes in this pathway |
| RODRIGUES NTN1 TARGETS DN | 158 | 102 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS UP | 214 | 155 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA HIF1A DN | 110 | 78 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 240 HELA | 60 | 43 | All SZGR 2.0 genes in this pathway |
| LENAOUR DENDRITIC CELL MATURATION UP | 114 | 84 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C5 | 46 | 36 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL DN | 216 | 143 | All SZGR 2.0 genes in this pathway |
| ALONSO METASTASIS UP | 198 | 128 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S3 | 266 | 180 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES DN | 210 | 141 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS NOT VIA AKT1 UP | 211 | 131 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS VIA AKT1 UP | 281 | 183 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS DN | 315 | 215 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 7 | 403 | 240 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38 | 337 | 236 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |