Gene Page: SPP2
Summary ?
| GeneID | 6694 |
| Symbol | SPP2 |
| Synonyms | SPP-24|SPP24 |
| Description | secreted phosphoprotein 2 |
| Reference | MIM:602637|HGNC:HGNC:11256|Ensembl:ENSG00000072080|HPRD:04029|Vega:OTTHUMG00000059208 |
| Gene type | protein-coding |
| Map location | 2q37.1 |
| Pascal p-value | 0.198 |
| eGene | Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
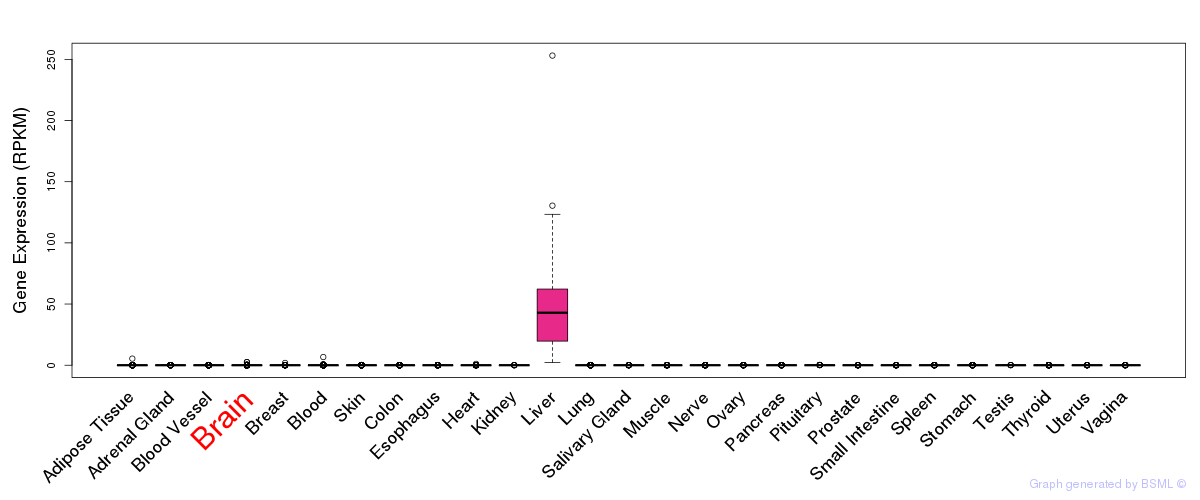
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NFE2L1 | 0.90 | 0.89 |
| ATG9A | 0.89 | 0.86 |
| DYNLL2 | 0.89 | 0.89 |
| MPP1 | 0.89 | 0.85 |
| WDR42A | 0.88 | 0.87 |
| KIAA0319L | 0.88 | 0.86 |
| SLC7A8 | 0.88 | 0.91 |
| DCTN1 | 0.88 | 0.89 |
| TOM1L2 | 0.88 | 0.86 |
| AC135048.1 | 0.87 | 0.85 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.45 | -0.29 |
| FAM159B | -0.43 | -0.58 |
| RPL35 | -0.41 | -0.47 |
| RPL31 | -0.40 | -0.43 |
| GNG11 | -0.39 | -0.31 |
| IL32 | -0.39 | -0.32 |
| C1orf54 | -0.39 | -0.31 |
| C1orf61 | -0.39 | -0.34 |
| RPS20 | -0.38 | -0.50 |
| RPL13AP22 | -0.38 | -0.54 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ZHOU INFLAMMATORY RESPONSE LIVE UP | 485 | 293 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA UP | 544 | 308 | All SZGR 2.0 genes in this pathway |
| HSIAO LIVER SPECIFIC GENES | 244 | 154 | All SZGR 2.0 genes in this pathway |
| SU LIVER | 55 | 32 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE DN | 274 | 165 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K27ME3 UP | 295 | 149 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT DN | 222 | 141 | All SZGR 2.0 genes in this pathway |
| OHGUCHI LIVER HNF4A TARGETS DN | 149 | 85 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| VANDESLUIS COMMD1 TARGETS GROUP 3 UP | 89 | 50 | All SZGR 2.0 genes in this pathway |