Gene Page: SRP68
Summary ?
| GeneID | 6730 |
| Symbol | SRP68 |
| Synonyms | - |
| Description | signal recognition particle 68kDa |
| Reference | MIM:604858|HGNC:HGNC:11302|Ensembl:ENSG00000167881|HPRD:05331|Vega:OTTHUMG00000180054 |
| Gene type | protein-coding |
| Map location | 17q25.1 |
| Pascal p-value | 0.172 |
| Sherlock p-value | 0.18 |
| Fetal beta | -0.754 |
| DMG | 1 (# studies) |
| eGene | Cerebellum Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg22467534 | 17 | 73841658 | SRP68 | 1.77E-6 | 4.294 | DMG:vanEijk_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs346790 | chr17 | 74300294 | SRP68 | 6730 | 0.16 | cis |
Section II. Transcriptome annotation
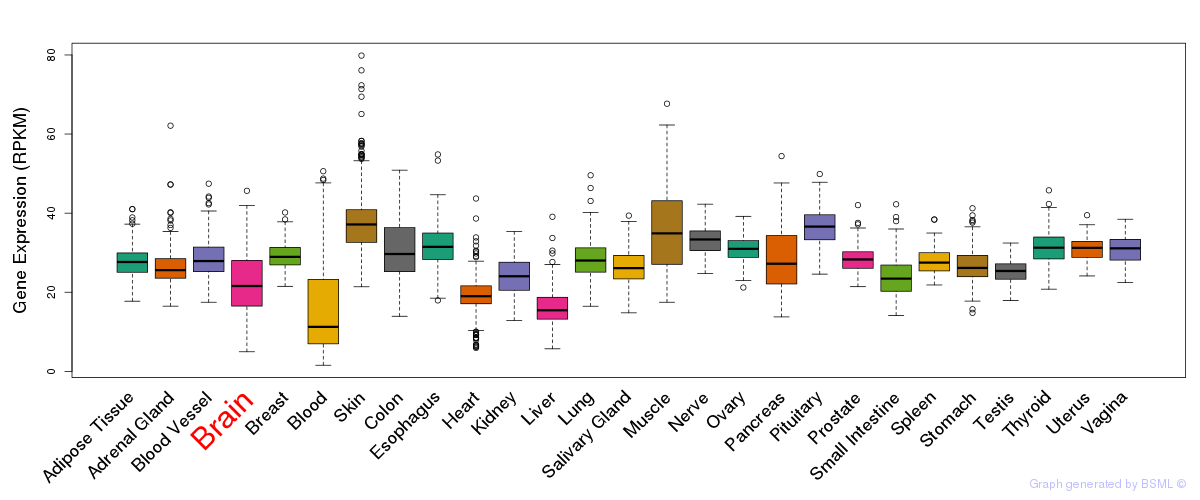
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| OGDHL | 0.90 | 0.85 |
| CPEB1 | 0.89 | 0.85 |
| FADS6 | 0.88 | 0.88 |
| RCAN2 | 0.87 | 0.91 |
| EHD3 | 0.87 | 0.86 |
| CNTNAP1 | 0.86 | 0.88 |
| RAB11FIP5 | 0.85 | 0.80 |
| KCNAB2 | 0.85 | 0.88 |
| GOT1 | 0.85 | 0.89 |
| LYNX1 | 0.85 | 0.81 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KIAA1949 | -0.49 | -0.40 |
| TUBB2B | -0.48 | -0.50 |
| SH3BP2 | -0.48 | -0.52 |
| RBMX2 | -0.48 | -0.51 |
| PDE9A | -0.47 | -0.48 |
| BCL7C | -0.47 | -0.55 |
| TRAF4 | -0.47 | -0.50 |
| ZNF300 | -0.46 | -0.27 |
| C9orf46 | -0.46 | -0.49 |
| ZNF311 | -0.46 | -0.32 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PROTEIN EXPORT | 24 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSLATION | 222 | 75 | All SZGR 2.0 genes in this pathway |
| REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | 179 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF PROTEINS | 518 | 242 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS DN | 352 | 225 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER UP | 181 | 108 | All SZGR 2.0 genes in this pathway |
| PASQUALUCCI LYMPHOMA BY GC STAGE UP | 283 | 177 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 17Q21 Q25 AMPLICON | 335 | 181 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 2 | 473 | 224 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN | 270 | 181 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |