Gene Page: SSTR3
Summary ?
| GeneID | 6753 |
| Symbol | SSTR3 |
| Synonyms | SS-3-R|SS3-R|SS3R|SSR-28 |
| Description | somatostatin receptor 3 |
| Reference | MIM:182453|HGNC:HGNC:11332|Ensembl:ENSG00000278195|HPRD:01675|Vega:OTTHUMG00000150537 |
| Gene type | protein-coding |
| Map location | 22q13.1 |
| Pascal p-value | 0.052 |
| Fetal beta | 0.293 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0471 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg14757173 | 22 | 37602542 | SSTR3 | 4.61E-5 | -0.262 | 0.021 | DMG:Wockner_2014 |
| cg03389133 | 22 | 37608611 | SSTR3 | 2.527E-4 | -0.229 | 0.037 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
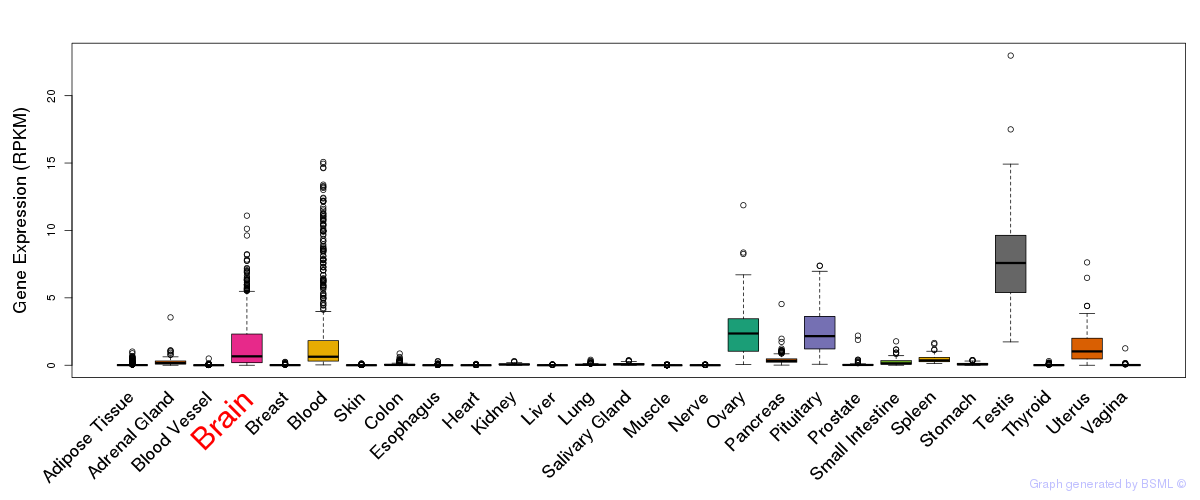
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PHF12 | 0.82 | 0.83 |
| ST3GAL1 | 0.82 | 0.78 |
| BAZ2A | 0.82 | 0.79 |
| EP300 | 0.82 | 0.81 |
| MICALL1 | 0.82 | 0.84 |
| SMG6 | 0.82 | 0.82 |
| SEPN1 | 0.82 | 0.88 |
| KIAA0355 | 0.81 | 0.84 |
| GATAD2A | 0.81 | 0.81 |
| PIK3C2B | 0.81 | 0.82 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.63 | -0.69 |
| ACOT13 | -0.61 | -0.67 |
| ACYP2 | -0.57 | -0.62 |
| MYL3 | -0.56 | -0.66 |
| AF347015.31 | -0.56 | -0.69 |
| CA4 | -0.55 | -0.60 |
| AF347015.27 | -0.55 | -0.66 |
| MT3 | -0.55 | -0.66 |
| B2M | -0.55 | -0.62 |
| S100B | -0.54 | -0.63 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004872 | receptor activity | IEA | - | |
| GO:0004994 | somatostatin receptor activity | TAS | 8405411 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007187 | G-protein signaling, coupled to cyclic nucleotide second messenger | TAS | 8405411 | |
| GO:0007267 | cell-cell signaling | TAS | 1337145 | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0008285 | negative regulation of cell proliferation | TAS | 8961277 | |
| GO:0008628 | induction of apoptosis by hormones | TAS | 8961277 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 1337145 | |
| GO:0031513 | nonmotile primary cilium | ISS | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME PEPTIDE LIGAND BINDING RECEPTORS | 188 | 108 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | 305 | 177 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA I SIGNALLING EVENTS | 195 | 114 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR DN | 37 | 20 | All SZGR 2.0 genes in this pathway |