Gene Page: TEK
Summary ?
| GeneID | 7010 |
| Symbol | TEK |
| Synonyms | CD202B|TIE-2|TIE2|VMCM|VMCM1 |
| Description | TEK receptor tyrosine kinase |
| Reference | MIM:600221|HGNC:HGNC:11724|Ensembl:ENSG00000120156|HPRD:02571|Vega:OTTHUMG00000019712 |
| Gene type | protein-coding |
| Map location | 9p21 |
| Pascal p-value | 1.501E-4 |
| Sherlock p-value | 0.103 |
| TADA p-value | 0.003 |
| Fetal beta | -1.312 |
| eGene | Cerebellar Hemisphere Cerebellum Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:McCarthy_2014 | Whole Exome Sequencing analysis | Whole exome sequencing of 57 trios with sporadic or familial schizophrenia. | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| TEK | chr9 | 27197558 | G | T | NM_000459 | p.E624X | nonsense SNV | C | NA | Schizophrenia | DNM:McCarthy_2014 |
Section II. Transcriptome annotation
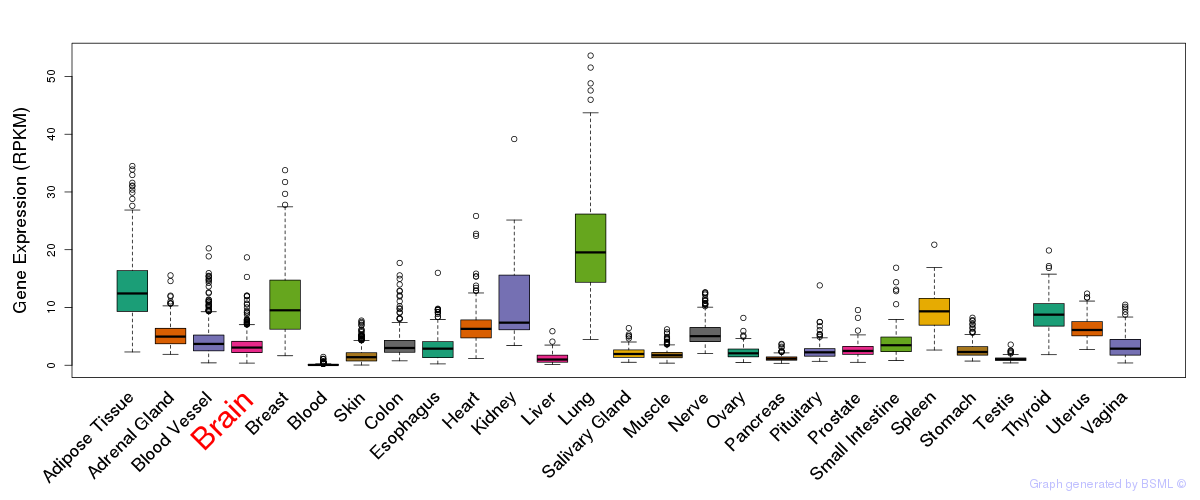
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HLA-E | 0.83 | 0.84 |
| LRRC32 | 0.83 | 0.90 |
| MMRN2 | 0.83 | 0.85 |
| PTRF | 0.81 | 0.87 |
| EPAS1 | 0.81 | 0.87 |
| ABCB1 | 0.81 | 0.85 |
| PRELP | 0.80 | 0.85 |
| CLEC14A | 0.78 | 0.83 |
| APOL3 | 0.77 | 0.73 |
| TNS1 | 0.77 | 0.84 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FRG1 | -0.60 | -0.67 |
| POLB | -0.59 | -0.67 |
| C18orf21 | -0.58 | -0.63 |
| MED19 | -0.58 | -0.61 |
| TCTEX1D2 | -0.58 | -0.67 |
| COMMD3 | -0.57 | -0.66 |
| SCNM1 | -0.56 | -0.64 |
| OXSM | -0.56 | -0.66 |
| ZNF32 | -0.56 | -0.68 |
| CWC15 | -0.56 | -0.63 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004714 | transmembrane receptor protein tyrosine kinase activity | TAS | neurite (GO term level: 8) | 8382358 |
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0004872 | receptor activity | IEA | - | |
| GO:0004872 | receptor activity | TAS | 10766762 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004713 | protein tyrosine kinase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0016301 | kinase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | TAS | 8980225 | |
| GO:0007160 | cell-matrix adhesion | IEA | - | |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0007267 | cell-cell signaling | TAS | 8980225 | |
| GO:0007165 | signal transduction | TAS | 8382358 | |
| GO:0016337 | cell-cell adhesion | IEA | - | |
| GO:0030334 | regulation of cell migration | IEA | - | |
| GO:0045765 | regulation of angiogenesis | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | EXP | 14665640 |14749497 | |
| GO:0005887 | integral to plasma membrane | TAS | 8382358 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ANGPT1 | AGP1 | AGPT | ANG1 | angiopoietin 1 | - | HPRD | 8980223 |8980224 |9723709 |12427764 |
| ANGPT1 | AGP1 | AGPT | ANG1 | angiopoietin 1 | Tie2 interacts with Ang1. | BIND | 15542434 |
| ANGPT1 | AGP1 | AGPT | ANG1 | angiopoietin 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 8980223 |9723709 |12427764 |
| ANGPT2 | AGPT2 | ANG2 | angiopoietin 2 | The receptor binding domain (RBD) of Ang2 interacts with the Tie2 ligand binding domain (LBD). This interaction was modelled on a demonstrated interaction between human Ang2 and Tie2 from an unspecified species. | BIND | 15893672 |
| ANGPT2 | AGPT2 | ANG2 | angiopoietin 2 | Tie2 interacts with Ang2. | BIND | 15542434 |
| ANGPT2 | AGPT2 | ANG2 | angiopoietin 2 | - | HPRD,BioGRID | 9723709 |
| ANGPT4 | AGP4 | ANG-3 | ANG4 | MGC138181 | MGC138183 | angiopoietin 4 | Tie2 interacts with Ang4. | BIND | 15542434 |
| ANGPT4 | AGP4 | ANG-3 | ANG4 | MGC138181 | MGC138183 | angiopoietin 4 | - | HPRD,BioGRID | 10051567 |
| ANGPTL1 | ANG3 | ANGPT3 | ARP1 | AngY | KIAA0351 | UNQ162 | dJ595C2.2 | angiopoietin-like 1 | Tie2 interacts with Ang3. This interaction was modeled on a demonstrated interaction between human Tie2 and mouse Ang3. | BIND | 15542434 |
| ANGPTL1 | ANG3 | ANGPT3 | ARP1 | AngY | KIAA0351 | UNQ162 | dJ595C2.2 | angiopoietin-like 1 | - | HPRD | 10051567 |
| DOK2 | p56DOK | p56dok-2 | docking protein 2, 56kDa | - | HPRD,BioGRID | 9764820 |
| GRB14 | - | growth factor receptor-bound protein 14 | - | HPRD | 10521483 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | - | HPRD | 7478529 |10521483 |
| GRB7 | - | growth factor receptor-bound protein 7 | - | HPRD | 10521483 |
| LCK | YT16 | p56lck | pp58lck | lymphocyte-specific protein tyrosine kinase | - | HPRD | 7545683 |
| PIK3R1 | GRB1 | p85 | p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | - | HPRD | 10521483 |
| PTPN11 | BPTP3 | CFC | MGC14433 | NS1 | PTP-1D | PTP2C | SH-PTP2 | SH-PTP3 | SHP2 | protein tyrosine phosphatase, non-receptor type 11 | - | HPRD | 10521483 |
| SOCS1 | CIS1 | CISH1 | JAB | SOCS-1 | SSI-1 | SSI1 | TIP3 | suppressor of cytokine signaling 1 | Socs1 interacts with TEK. This interaction was modeled on a demonstrated interaction between mouse Socs1 and Tek from an unspecified species. | BIND | 10022833 |
| SOCS1 | CIS1 | CISH1 | JAB | SOCS-1 | SSI-1 | SSI1 | TIP3 | suppressor of cytokine signaling 1 | - | HPRD | 10022833 |
| TIE1 | JTK14 | TIE | tyrosine kinase with immunoglobulin-like and EGF-like domains 1 | - | HPRD,BioGRID | 10995770 |
| TNIP2 | ABIN-2 | ABIN2 | FLIP1 | KLIP | MGC4289 | TNFAIP3 interacting protein 2 | - | HPRD | 12609966 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID ANGIOPOIETIN RECEPTOR PATHWAY | 50 | 41 | All SZGR 2.0 genes in this pathway |
| PID SHP2 PATHWAY | 58 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | 91 | 65 | All SZGR 2.0 genes in this pathway |
| REACTOME TIE2 SIGNALING | 17 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER DN | 232 | 154 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS F UP | 185 | 119 | All SZGR 2.0 genes in this pathway |
| TAKADA GASTRIC CANCER COPY NUMBER DN | 29 | 15 | All SZGR 2.0 genes in this pathway |
| INGRAM SHH TARGETS UP | 127 | 79 | All SZGR 2.0 genes in this pathway |
| BERTUCCI INVASIVE CARCINOMA DUCTAL VS LOBULAR DN | 46 | 34 | All SZGR 2.0 genes in this pathway |
| ROSS AML OF FAB M7 TYPE | 68 | 44 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 1 UP | 182 | 119 | All SZGR 2.0 genes in this pathway |
| HARRIS HYPOXIA | 81 | 64 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY UP | 86 | 48 | All SZGR 2.0 genes in this pathway |
| ZHANG PROLIFERATING VS QUIESCENT | 51 | 41 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| KRASNOSELSKAYA ILF3 TARGETS DN | 46 | 38 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN DN | 353 | 226 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL DN | 216 | 143 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA METAGENE | 242 | 168 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS UP | 165 | 118 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER LATE RECURRENCE DN | 69 | 48 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 1 UP | 125 | 61 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 3 UP | 170 | 97 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 6 UP | 140 | 81 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS UP | 387 | 225 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| WIEDERSCHAIN TARGETS OF BMI1 AND PCGF2 | 57 | 39 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| MADAN DPPA4 TARGETS | 46 | 26 | All SZGR 2.0 genes in this pathway |