Gene Page: TGFB1
Summary ?
| GeneID | 7040 |
| Symbol | TGFB1 |
| Synonyms | CED|DPD1|LAP|TGFB|TGFbeta |
| Description | transforming growth factor beta 1 |
| Reference | MIM:190180|HGNC:HGNC:11766|Ensembl:ENSG00000105329|HPRD:01821|Vega:OTTHUMG00000182760 |
| Gene type | protein-coding |
| Map location | 19q13.1 |
| Pascal p-value | 0.065 |
| Fetal beta | -0.566 |
| eGene | Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0382 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
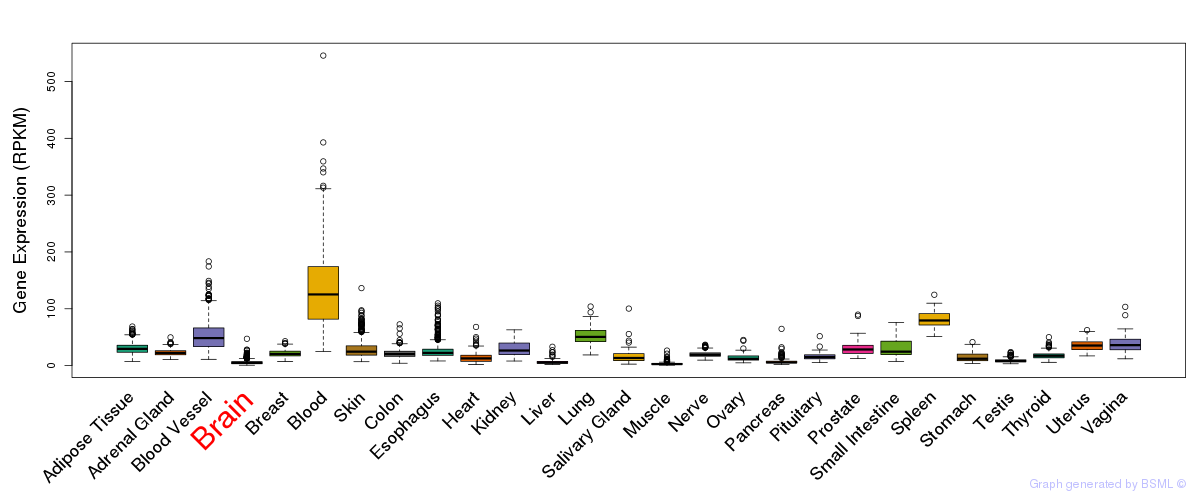
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CENPM | 0.91 | 0.61 |
| CDC45L | 0.90 | 0.59 |
| MCM5 | 0.90 | 0.67 |
| FAM83D | 0.89 | 0.61 |
| AC087645.2 | 0.89 | 0.64 |
| PCNA | 0.89 | 0.73 |
| TACC3 | 0.89 | 0.63 |
| ASF1B | 0.88 | 0.62 |
| PLK1 | 0.88 | 0.65 |
| FAM64A | 0.88 | 0.63 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.39 | -0.59 |
| AF347015.27 | -0.39 | -0.61 |
| MT-CO2 | -0.38 | -0.60 |
| C5orf53 | -0.38 | -0.44 |
| AF347015.33 | -0.38 | -0.61 |
| AF347015.8 | -0.38 | -0.62 |
| AF347015.15 | -0.38 | -0.62 |
| MT-CYB | -0.37 | -0.60 |
| AIFM3 | -0.37 | -0.46 |
| AF347015.2 | -0.36 | -0.61 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005114 | type II transforming growth factor beta receptor binding | IDA | 11157754 | |
| GO:0005515 | protein binding | ISS | - | |
| GO:0008083 | growth factor activity | IEA | - | |
| GO:0016563 | transcription activator activity | ISS | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0043011 | myeloid dendritic cell differentiation | IEA | dendrite (GO term level: 10) | - |
| GO:0001501 | skeletal system development | ISS | - | |
| GO:0002248 | connective tissue replacement during inflammatory response | TAS | 9639571 | |
| GO:0002513 | tolerance induction to self antigen | IEA | - | |
| GO:0001837 | epithelial to mesenchymal transition | TAS | 14679171 | |
| GO:0007184 | SMAD protein nuclear translocation | IDA | 15334054 | |
| GO:0006468 | protein amino acid phosphorylation | ISS | - | |
| GO:0008219 | cell death | ISS | - | |
| GO:0006917 | induction of apoptosis | IDA | 15334054 | |
| GO:0009887 | organ morphogenesis | ISS | - | |
| GO:0007050 | cell cycle arrest | IDA | 14555988 | |
| GO:0008284 | positive regulation of cell proliferation | ISS | - | |
| GO:0008354 | germ cell migration | IEA | - | |
| GO:0007435 | salivary gland morphogenesis | IEP | 18080134 | |
| GO:0016202 | regulation of striated muscle development | ISS | - | |
| GO:0006796 | phosphate metabolic process | IDA | 10513816 | |
| GO:0006754 | ATP biosynthetic process | IDA | 10513816 | |
| GO:0006954 | inflammatory response | ISS | - | |
| GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway | IDA | 15334054 | |
| GO:0019049 | evasion of host defenses by virus | IDA | 15334054 | |
| GO:0042306 | regulation of protein import into nucleus | ISS | - | |
| GO:0016481 | negative regulation of transcription | IDA | 15702480 | |
| GO:0051101 | regulation of DNA binding | ISS | - | |
| GO:0042130 | negative regulation of T cell proliferation | IEA | - | |
| GO:0030501 | positive regulation of bone mineralization | IEP | 17401695 | |
| GO:0048298 | positive regulation of isotype switching to IgA isotypes | IDA | 14988498 | |
| GO:0030879 | mammary gland development | IEA | - | |
| GO:0032740 | positive regulation of interleukin-17 production | IDA | 18453574 | |
| GO:0031575 | G1/S transition checkpoint | IDA | 15334054 | |
| GO:0030279 | negative regulation of ossification | IEA | - | |
| GO:0032943 | mononuclear cell proliferation | IEA | - | |
| GO:0032967 | positive regulation of collagen biosynthetic process | TAS | 9639571 | |
| GO:0030308 | negative regulation of cell growth | IDA | 15334054 | |
| GO:0035307 | positive regulation of protein amino acid dephosphorylation | IDA | 14555988 | |
| GO:0043029 | T cell homeostasis | IEA | - | |
| GO:0045893 | positive regulation of transcription, DNA-dependent | ISS | - | |
| GO:0045930 | negative regulation of mitotic cell cycle | IDA | 14555988 | |
| GO:0050680 | negative regulation of epithelial cell proliferation | IMP | 16943770 | |
| GO:0045066 | regulatory T cell differentiation | IEA | - | |
| GO:0043932 | ossification involved in bone remodeling | IEP | 17401695 | |
| GO:0048535 | lymph node development | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005796 | Golgi lumen | EXP | 3457014 |9934696 | |
| GO:0005576 | extracellular region | EXP | 9865696 |9934696 |11100470 | |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005578 | proteinaceous extracellular matrix | ISS | - | |
| GO:0005615 | extracellular space | ISS | - | |
| GO:0005634 | nucleus | IDA | 18040277 | |
| GO:0005737 | cytoplasm | IDA | 18040277 | |
| GO:0031093 | platelet alpha granule lumen | EXP | 3457014 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACVRL1 | ACVRLK1 | ALK-1 | ALK1 | HHT | HHT2 | ORW2 | SKR3 | TSR-I | activin A receptor type II-like 1 | - | HPRD,BioGRID | 10716993 |
| APP | AAA | ABETA | ABPP | AD1 | APPI | CTFgamma | CVAP | PN2 | amyloid beta (A4) precursor protein | TGF-beta-1 interacts with Amyloid-beta. This interaction was modelled on a demonstrated interaction between TGF-beta-1 from an unspecified species and consensus Amyloid-beta peptides. | BIND | 12867422 |
| BGN | DSPG1 | PG-S1 | PGI | SLRR1A | biglycan | - | HPRD,BioGRID | 8093006 |10383378 |
| BMP2 | BMP2A | bone morphogenetic protein 2 | - | HPRD | 11456401 |
| BMP3 | BMP-3A | bone morphogenetic protein 3 | - | HPRD | 9434787 |
| CCL3 | G0S19-1 | LD78ALPHA | MIP-1-alpha | MIP1A | SCYA3 | chemokine (C-C motif) ligand 3 | - | HPRD | 8568261 |
| COL2A1 | ANFH | AOM | COL11A3 | MGC131516 | SEDC | collagen, type II, alpha 1 | - | HPRD,BioGRID | 10085302 |
| CTGF | CCN2 | HCS24 | IGFBP8 | MGC102839 | NOV2 | connective tissue growth factor | - | HPRD,BioGRID | 10457363 |
| DAXX | BING2 | DAP6 | EAP1 | MGC126245 | MGC126246 | death-domain associated protein | - | HPRD | 11483955 |
| DCN | CSCD | DSPG2 | PG40 | PGII | PGS2 | SLRR1B | decorin | - | HPRD | 7638106 |7798269 |
| DCN | CSCD | DSPG2 | PG40 | PGII | PGS2 | SLRR1B | decorin | Reconstituted Complex | BioGRID | 7798269 |8093006 |9675033 |
| EIF3I | EIF3S2 | PRO2242 | TRIP-1 | TRIP1 | eIF3-beta | eIF3-p36 | eukaryotic translation initiation factor 3, subunit I | - | HPRD,BioGRID | 9813058 |
| ENG | CD105 | END | FLJ41744 | HHT1 | ORW | ORW1 | endoglin | - | HPRD | 12015308 |
| FCN1 | FCNM | ficolin (collagen/fibrinogen domain containing) 1 | - | HPRD,BioGRID | 7686157 |
| FMOD | SLRR2E | fibromodulin | - | HPRD,BioGRID | 8093006 |
| FNTA | FPTA | MGC99680 | PGGT1A | PTAR2 | farnesyltransferase, CAAX box, alpha | - | HPRD,BioGRID | 8599089 |
| ITGAV | CD51 | DKFZp686A08142 | MSK8 | VNRA | integrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51) | - | HPRD | 11970960 |
| ITGB6 | - | integrin, beta 6 | - | HPRD | 10025398 |
| ITGB8 | - | integrin, beta 8 | - | HPRD | 11970960 |
| LTBP1 | MGC163161 | latent transforming growth factor beta binding protein 1 | - | HPRD,BioGRID | 8617200 |10930463 |
| LTBP3 | DKFZp586M2123 | FLJ33431 | FLJ39893 | FLJ42533 | FLJ44138 | LTBP-3 | LTBP2 | pp6425 | latent transforming growth factor beta binding protein 3 | - | HPRD | 12062452 |
| LTBP4 | FLJ46318 | FLJ90018 | LTBP-4 | LTBP-4L | latent transforming growth factor beta binding protein 4 | - | HPRD | 9660815 |
| MMP2 | CLG4 | CLG4A | MMP-II | MONA | TBE-1 | matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) | - | HPRD,BioGRID | 10652271 |
| MMP9 | CLG4B | GELB | MMP-9 | matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) | Biochemical Activity | BioGRID | 10652271 |
| PZP | CPAMD6 | MGC133093 | pregnancy-zone protein | - | HPRD,BioGRID | 7513640 |
| SNIP1 | FLJ12553 | RP3-423B22.3 | dJ423B22.2 | Smad nuclear interacting protein 1 | Affinity Capture-Western | BioGRID | 10887155 |
| SPARC | ON | secreted protein, acidic, cysteine-rich (osteonectin) | - | HPRD | 15034927 |
| TGFB1 | CED | DPD1 | TGFB | TGFbeta | transforming growth factor, beta 1 | - | HPRD | 7935686 |
| TGFB2 | MGC116892 | TGF-beta2 | transforming growth factor, beta 2 | Two-hybrid | BioGRID | 11483955 |
| TGFB2 | MGC116892 | TGF-beta2 | transforming growth factor, beta 2 | - | HPRD | 11157754 |
| TGFBR1 | AAT5 | ACVRLK4 | ALK-5 | ALK5 | LDS1A | LDS2A | SKR4 | TGFR-1 | transforming growth factor, beta receptor 1 | TGF-beta-1 interacts with TGF-beta-R-I. This interaction was modeled on a demonstrated interaction between TGF-beta-1 from an unspecified species and human TGF-beta-R-I. | BIND | 7852346 |
| TGFBR1 | AAT5 | ACVRLK4 | ALK-5 | ALK5 | LDS1A | LDS2A | SKR4 | TGFR-1 | transforming growth factor, beta receptor 1 | - | HPRD,BioGRID | 8235612 |
| TGFBR2 | AAT3 | FAA3 | LDS1B | LDS2B | MFS2 | RIIC | TAAD2 | TGFR-2 | TGFbeta-RII | transforming growth factor, beta receptor II (70/80kDa) | Affinity Capture-Western | BioGRID | 11157754 |
| TGFBR2 | AAT3 | FAA3 | LDS1B | LDS2B | MFS2 | RIIC | TAAD2 | TGFR-2 | TGFbeta-RII | transforming growth factor, beta receptor II (70/80kDa) | - | HPRD | 7935686 |
| TGFBR2 | AAT3 | FAA3 | LDS1B | LDS2B | MFS2 | RIIC | TAAD2 | TGFR-2 | TGFbeta-RII | transforming growth factor, beta receptor II (70/80kDa) | TGF-beta-1 interacts with TGF-beta-R-II. This interaction was modeled on a demonstrated interaction between TGF-beta-1 from an unspecified species and human TGF-beta-R-II. | BIND | 7852346 |
| TGFBR3 | BGCAN | betaglycan | transforming growth factor, beta receptor III | - | HPRD | 7935686 |
| TGFBR3 | BGCAN | betaglycan | transforming growth factor, beta receptor III | Affinity Capture-Western | BioGRID | 8106553 |
| TGFBRAP1 | TRAP-1 | TRAP1 | transforming growth factor, beta receptor associated protein 1 | Affinity Capture-Western | BioGRID | 11278302 |
| THBS1 | THBS | TSP | TSP1 | thrombospondin 1 | - | HPRD,BioGRID | 1550960 |
| VASN | SLITL2 | vasorin | - | HPRD,BioGRID | 15247411 |
| VTN | V75 | VN | VNT | vitronectin | - | HPRD,BioGRID | 11796824 |
| YWHAE | 14-3-3E | FLJ45465 | KCIP-1 | MDCR | MDS | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | - | HPRD,BioGRID | 11172812 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION | 267 | 161 | All SZGR 2.0 genes in this pathway |
| KEGG CELL CYCLE | 128 | 84 | All SZGR 2.0 genes in this pathway |
| KEGG TGF BETA SIGNALING PATHWAY | 86 | 64 | All SZGR 2.0 genes in this pathway |
| KEGG INTESTINAL IMMUNE NETWORK FOR IGA PRODUCTION | 48 | 36 | All SZGR 2.0 genes in this pathway |
| KEGG LEISHMANIA INFECTION | 72 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG COLORECTAL CANCER | 62 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG RENAL CELL CARCINOMA | 70 | 60 | All SZGR 2.0 genes in this pathway |
| KEGG PANCREATIC CANCER | 70 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG CHRONIC MYELOID LEUKEMIA | 73 | 59 | All SZGR 2.0 genes in this pathway |
| KEGG HYPERTROPHIC CARDIOMYOPATHY HCM | 85 | 65 | All SZGR 2.0 genes in this pathway |
| KEGG DILATED CARDIOMYOPATHY | 92 | 68 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ALK PATHWAY | 37 | 29 | All SZGR 2.0 genes in this pathway |
| BIOCARTA G1 PATHWAY | 28 | 21 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CTCF PATHWAY | 23 | 18 | All SZGR 2.0 genes in this pathway |
| BIOCARTA INFLAM PATHWAY | 29 | 24 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ERYTH PATHWAY | 15 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MAPK PATHWAY | 87 | 68 | All SZGR 2.0 genes in this pathway |
| BIOCARTA P38MAPK PATHWAY | 40 | 31 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TOB1 PATHWAY | 21 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NKT PATHWAY | 29 | 21 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL1R PATHWAY | 33 | 29 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TGFB PATHWAY | 19 | 14 | All SZGR 2.0 genes in this pathway |
| SA MMP CYTOKINE CONNECTION | 15 | 9 | All SZGR 2.0 genes in this pathway |
| PID GLYPICAN 1PATHWAY | 27 | 20 | All SZGR 2.0 genes in this pathway |
| PID IL27 PATHWAY | 26 | 20 | All SZGR 2.0 genes in this pathway |
| PID TELOMERASE PATHWAY | 68 | 48 | All SZGR 2.0 genes in this pathway |
| PID RXR VDR PATHWAY | 26 | 24 | All SZGR 2.0 genes in this pathway |
| PID AP1 PATHWAY | 70 | 60 | All SZGR 2.0 genes in this pathway |
| PID UPA UPAR PATHWAY | 42 | 30 | All SZGR 2.0 genes in this pathway |
| PID ALK1 PATHWAY | 26 | 21 | All SZGR 2.0 genes in this pathway |
| PID SYNDECAN 1 PATHWAY | 46 | 29 | All SZGR 2.0 genes in this pathway |
| PID SYNDECAN 2 PATHWAY | 33 | 27 | All SZGR 2.0 genes in this pathway |
| PID TGFBR PATHWAY | 55 | 38 | All SZGR 2.0 genes in this pathway |
| PID IL12 STAT4 PATHWAY | 33 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | 16 | 7 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | 23 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | 26 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | 89 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | 72 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY TGF BETA RECEPTOR COMPLEX | 63 | 42 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA UP | 305 | 185 | All SZGR 2.0 genes in this pathway |
| SMIRNOV CIRCULATING ENDOTHELIOCYTES IN CANCER UP | 158 | 103 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION KRAS UP | 126 | 72 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2A DN | 141 | 84 | All SZGR 2.0 genes in this pathway |
| BARIS THYROID CANCER DN | 59 | 45 | All SZGR 2.0 genes in this pathway |
| STREICHER LSM1 TARGETS DN | 19 | 13 | All SZGR 2.0 genes in this pathway |
| RAMJAUN APOPTOSIS BY TGFB1 VIA SMAD4 UP | 7 | 6 | All SZGR 2.0 genes in this pathway |
| BREUHAHN GROWTH FACTOR SIGNALING IN LIVER CANCER | 22 | 19 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| KHETCHOUMIAN TRIM24 TARGETS UP | 47 | 38 | All SZGR 2.0 genes in this pathway |
| GALIE TUMOR ANGIOGENESIS | 8 | 7 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| SHIN B CELL LYMPHOMA CLUSTER 3 | 28 | 23 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 1 UP | 140 | 85 | All SZGR 2.0 genes in this pathway |
| LAMB CCND1 TARGETS | 19 | 14 | All SZGR 2.0 genes in this pathway |
| PENG LEUCINE DEPRIVATION DN | 187 | 122 | All SZGR 2.0 genes in this pathway |
| TENEDINI MEGAKARYOCYTE MARKERS | 66 | 48 | All SZGR 2.0 genes in this pathway |
| FERNANDEZ BOUND BY MYC | 182 | 116 | All SZGR 2.0 genes in this pathway |
| NING CHRONIC OBSTRUCTIVE PULMONARY DISEASE UP | 157 | 105 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION DN | 337 | 230 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS DN | 366 | 238 | All SZGR 2.0 genes in this pathway |
| NEMETH INFLAMMATORY RESPONSE LPS UP | 88 | 64 | All SZGR 2.0 genes in this pathway |
| KANG IMMORTALIZED BY TERT UP | 89 | 61 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND DN | 225 | 163 | All SZGR 2.0 genes in this pathway |
| ABRAHAM ALPC VS MULTIPLE MYELOMA DN | 19 | 14 | All SZGR 2.0 genes in this pathway |
| XU CREBBP TARGETS DN | 44 | 31 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| WU HBX TARGETS 2 DN | 16 | 11 | All SZGR 2.0 genes in this pathway |
| WU HBX TARGETS 1 DN | 23 | 14 | All SZGR 2.0 genes in this pathway |
| HARRIS HYPOXIA | 81 | 64 | All SZGR 2.0 genes in this pathway |
| MURAKAMI UV RESPONSE 6HR DN | 21 | 13 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP | 249 | 170 | All SZGR 2.0 genes in this pathway |
| CHIBA RESPONSE TO TSA UP | 52 | 33 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK UP | 244 | 151 | All SZGR 2.0 genes in this pathway |
| WU HBX TARGETS 3 DN | 13 | 9 | All SZGR 2.0 genes in this pathway |
| MOREIRA RESPONSE TO TSA UP | 28 | 23 | All SZGR 2.0 genes in this pathway |
| MURAKAMI UV RESPONSE 24HR | 20 | 13 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS UP | 280 | 183 | All SZGR 2.0 genes in this pathway |
| GAVIN PDE3B TARGETS | 22 | 15 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| MARZEC IL2 SIGNALING UP | 115 | 80 | All SZGR 2.0 genes in this pathway |
| POS HISTAMINE RESPONSE NETWORK | 32 | 22 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| GAUTSCHI SRC SIGNALING | 8 | 6 | All SZGR 2.0 genes in this pathway |
| GAURNIER PSMD4 TARGETS | 73 | 55 | All SZGR 2.0 genes in this pathway |
| MATZUK CENTRAL FOR FEMALE FERTILITY | 29 | 25 | All SZGR 2.0 genes in this pathway |
| MATZUK SPERMATOZOA | 114 | 77 | All SZGR 2.0 genes in this pathway |
| LABBE WNT3A TARGETS UP | 112 | 71 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA UP | 259 | 185 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA METAGENE | 242 | 168 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS UP | 165 | 118 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS DN | 138 | 92 | All SZGR 2.0 genes in this pathway |
| GU PDEF TARGETS UP | 71 | 49 | All SZGR 2.0 genes in this pathway |
| HOFFMANN SMALL PRE BII TO IMMATURE B LYMPHOCYTE DN | 50 | 33 | All SZGR 2.0 genes in this pathway |
| HOFFMANN IMMATURE TO MATURE B LYMPHOCYTE DN | 50 | 36 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP | 395 | 249 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN DEDIFFERENTIATED STATE DN | 7 | 7 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE DN | 181 | 97 | All SZGR 2.0 genes in this pathway |
| WOO LIVER CANCER RECURRENCE UP | 105 | 75 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 AND H3K27ME3 | 349 | 234 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN | 253 | 192 | All SZGR 2.0 genes in this pathway |
| DANG MYC TARGETS DN | 31 | 25 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K4ME3 AND H3K27ME3 | 210 | 148 | All SZGR 2.0 genes in this pathway |
| DASU IL6 SIGNALING UP | 59 | 44 | All SZGR 2.0 genes in this pathway |
| KANG AR TARGETS DN | 19 | 12 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 UP | 140 | 94 | All SZGR 2.0 genes in this pathway |
| KASLER HDAC7 TARGETS 1 UP | 194 | 133 | All SZGR 2.0 genes in this pathway |
| JUBAN TARGETS OF SPI1 AND FLI1 DN | 92 | 60 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR UP | 199 | 143 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS | 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |