| KEGG TGF BETA SIGNALING PATHWAY
| 86 | 64 | All SZGR 2.0 genes in this pathway |
| KEGG FOCAL ADHESION
| 201 | 138 | All SZGR 2.0 genes in this pathway |
| KEGG ECM RECEPTOR INTERACTION
| 84 | 53 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN1 PATHWAY
| 66 | 44 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN A4B1 PATHWAY
| 33 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY PDGF
| 122 | 93 | All SZGR 2.0 genes in this pathway |
| WATANABE RECTAL CANCER RADIOTHERAPY RESPONSIVE DN
| 92 | 51 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE UP
| 351 | 230 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST LOBULAR CARCINOMA VS DUCTAL NORMAL UP
| 69 | 38 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST LOBULAR CARCINOMA VS LOBULAR NORMAL DN
| 74 | 42 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA UP
| 305 | 185 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER ADVANCED VS EARLY UP
| 175 | 120 | All SZGR 2.0 genes in this pathway |
| MOROSETTI FACIOSCAPULOHUMERAL MUSCULAR DISTROPHY UP
| 20 | 11 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN
| 1375 | 806 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS UP
| 293 | 179 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP
| 443 | 294 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION KRAS UP
| 126 | 72 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION CDC25 UP
| 120 | 73 | All SZGR 2.0 genes in this pathway |
| CHEBOTAEV GR TARGETS DN
| 120 | 73 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS A UP
| 191 | 128 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN
| 394 | 258 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK UP
| 271 | 175 | All SZGR 2.0 genes in this pathway |
| BEGUM TARGETS OF PAX3 FOXO1 FUSION DN
| 45 | 34 | All SZGR 2.0 genes in this pathway |
| HUMMERICH SKIN CANCER PROGRESSION UP
| 88 | 58 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 5
| 147 | 89 | All SZGR 2.0 genes in this pathway |
| LI AMPLIFIED IN LUNG CANCER
| 178 | 108 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B
| 392 | 251 | All SZGR 2.0 genes in this pathway |
| HERNANDEZ ABERRANT MITOSIS BY DOCETACEL 2NM UP
| 81 | 57 | All SZGR 2.0 genes in this pathway |
| HERNANDEZ MITOTIC ARREST BY DOCETAXEL 2 UP
| 64 | 45 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN
| 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN
| 483 | 336 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP
| 358 | 245 | All SZGR 2.0 genes in this pathway |
| HOWLIN PUBERTAL MAMMARY GLAND
| 69 | 40 | All SZGR 2.0 genes in this pathway |
| LIEN BREAST CARCINOMA METAPLASTIC
| 35 | 25 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS
| 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH OCT4 TARGETS
| 290 | 172 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS
| 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH NOS TARGETS
| 179 | 105 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3
| 1118 | 744 | All SZGR 2.0 genes in this pathway |
| SUNG METASTASIS STROMA UP
| 110 | 70 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS DN
| 234 | 137 | All SZGR 2.0 genes in this pathway |
| KANG IMMORTALIZED BY TERT DN
| 102 | 67 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C8
| 72 | 56 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN
| 408 | 274 | All SZGR 2.0 genes in this pathway |
| NIELSEN MALIGNAT FIBROUS HISTIOCYTOMA UP
| 18 | 11 | All SZGR 2.0 genes in this pathway |
| KUNINGER IGF1 VS PDGFB TARGETS DN
| 46 | 28 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN
| 504 | 323 | All SZGR 2.0 genes in this pathway |
| GERHOLD ADIPOGENESIS DN
| 64 | 44 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 24 HR DN
| 91 | 64 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA DELAYED RESPONSE TO TGFB1
| 39 | 26 | All SZGR 2.0 genes in this pathway |
| LEE AGING CEREBELLUM UP
| 84 | 58 | All SZGR 2.0 genes in this pathway |
| JACKSON DNMT1 TARGETS UP
| 77 | 57 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL DN
| 128 | 93 | All SZGR 2.0 genes in this pathway |
| LY AGING MIDDLE UP
| 14 | 11 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS PEAK AT 0HR
| 63 | 48 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA RESPONSE TO TGFB1 C4
| 13 | 10 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC UP
| 202 | 115 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 9
| 92 | 59 | All SZGR 2.0 genes in this pathway |
| URS ADIPOCYTE DIFFERENTIATION DN
| 30 | 20 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 12HR DN
| 101 | 64 | All SZGR 2.0 genes in this pathway |
| JI CARCINOGENESIS BY KRAS AND STK11 DN
| 17 | 12 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL UP
| 260 | 174 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA METAGENE
| 242 | 168 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS UP
| 165 | 118 | All SZGR 2.0 genes in this pathway |
| SEKI INFLAMMATORY RESPONSE LPS DN
| 23 | 16 | All SZGR 2.0 genes in this pathway |
| SCHRAETS MLL TARGETS DN
| 33 | 24 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK
| 1210 | 725 | All SZGR 2.0 genes in this pathway |
| CROONQUIST STROMAL STIMULATION UP
| 60 | 42 | All SZGR 2.0 genes in this pathway |
| CROONQUIST NRAS VS STROMAL STIMULATION DN
| 99 | 65 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G56 DN
| 17 | 9 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS CTNNB1 DN
| 170 | 105 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS ICP WITH H3K27ME3
| 54 | 32 | All SZGR 2.0 genes in this pathway |
| NAKAYAMA SOFT TISSUE TUMORS PCA1 UP
| 76 | 46 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS UP
| 221 | 135 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 2
| 86 | 50 | All SZGR 2.0 genes in this pathway |
| NAKAMURA ADIPOGENESIS EARLY DN
| 38 | 27 | All SZGR 2.0 genes in this pathway |
| NAKAMURA ADIPOGENESIS LATE DN
| 38 | 25 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT
| 572 | 352 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE
| 1137 | 655 | All SZGR 2.0 genes in this pathway |
| STEGER ADIPOGENESIS DN
| 25 | 15 | All SZGR 2.0 genes in this pathway |
| BILANGES SERUM SENSITIVE VIA TSC2
| 39 | 25 | All SZGR 2.0 genes in this pathway |
| SERVITJA ISLET HNF1A TARGETS UP
| 163 | 111 | All SZGR 2.0 genes in this pathway |
| WANG MLL TARGETS
| 289 | 188 | All SZGR 2.0 genes in this pathway |
| ANASTASSIOU CANCER MESENCHYMAL TRANSITION SIGNATURE
| 64 | 40 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY
| 1725 | 838 | All SZGR 2.0 genes in this pathway |
| NABA ECM GLYCOPROTEINS
| 196 | 99 | All SZGR 2.0 genes in this pathway |
| NABA CORE MATRISOME
| 275 | 148 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME
| 1028 | 559 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER CLUSTER 4
| 19 | 12 | All SZGR 2.0 genes in this pathway |
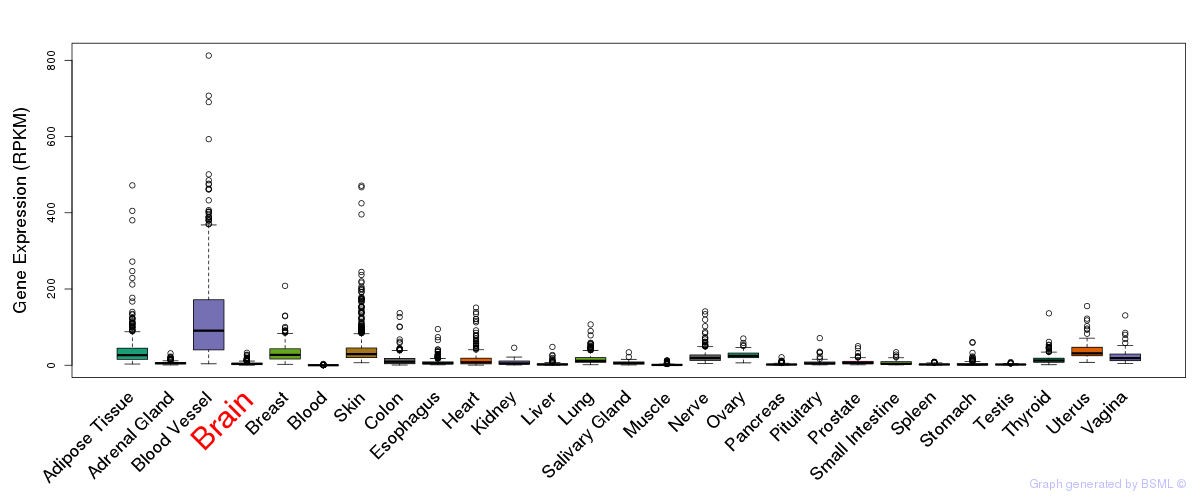
 Differentially methylated gene
Differentially methylated gene eQTL annotation
eQTL annotation