Gene Page: TRAF1
Summary ?
| GeneID | 7185 |
| Symbol | TRAF1 |
| Synonyms | EBI6|MGC:10353 |
| Description | TNF receptor associated factor 1 |
| Reference | MIM:601711|HGNC:HGNC:12031|Ensembl:ENSG00000056558|HPRD:03418| |
| Gene type | protein-coding |
| Map location | 9q33-q34 |
| Pascal p-value | 0.044 |
| Fetal beta | -0.098 |
| eGene | Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Hippocampus Hypothalamus Nucleus accumbens basal ganglia Putamen basal ganglia |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1953126 | 9 | 123640500 | TRAF1 | ENSG00000056558.6 | 1.636E-7 | 0 | 50951 | gtex_brain_putamen_basal |
| rs565526738 | 9 | 123641331 | TRAF1 | ENSG00000056558.6 | 3.377E-7 | 0 | 50120 | gtex_brain_putamen_basal |
| rs1930778 | 9 | 123641369 | TRAF1 | ENSG00000056558.6 | 1.804E-7 | 0 | 50082 | gtex_brain_putamen_basal |
| rs10760118 | 9 | 123641616 | TRAF1 | ENSG00000056558.6 | 3.377E-7 | 0 | 49835 | gtex_brain_putamen_basal |
| rs1609810 | 9 | 123642351 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 49100 | gtex_brain_putamen_basal |
| rs10818480 | 9 | 123643170 | TRAF1 | ENSG00000056558.6 | 1.375E-7 | 0 | 48281 | gtex_brain_putamen_basal |
| rs375675198 | 9 | 123643424 | TRAF1 | ENSG00000056558.6 | 2.356E-7 | 0 | 48027 | gtex_brain_putamen_basal |
| rs67054335 | 9 | 123643425 | TRAF1 | ENSG00000056558.6 | 3.113E-8 | 0 | 48026 | gtex_brain_putamen_basal |
| rs7034390 | 9 | 123646488 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 44963 | gtex_brain_putamen_basal |
| rs201783688 | 9 | 123646511 | TRAF1 | ENSG00000056558.6 | 1.274E-7 | 0 | 44940 | gtex_brain_putamen_basal |
| rs4323544 | 9 | 123647377 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 44074 | gtex_brain_putamen_basal |
| rs10739576 | 9 | 123647607 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 43844 | gtex_brain_putamen_basal |
| rs10760121 | 9 | 123647915 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 43536 | gtex_brain_putamen_basal |
| rs7868822 | 9 | 123650327 | TRAF1 | ENSG00000056558.6 | 5.695E-7 | 0 | 41124 | gtex_brain_putamen_basal |
| rs10818483 | 9 | 123650473 | TRAF1 | ENSG00000056558.6 | 1.034E-6 | 0 | 40978 | gtex_brain_putamen_basal |
| rs10760122 | 9 | 123650533 | TRAF1 | ENSG00000056558.6 | 2.102E-6 | 0 | 40918 | gtex_brain_putamen_basal |
| rs10760123 | 9 | 123650534 | TRAF1 | ENSG00000056558.6 | 1.759E-6 | 0 | 40917 | gtex_brain_putamen_basal |
| rs2270231 | 9 | 123650982 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 40469 | gtex_brain_putamen_basal |
| rs6478484 | 9 | 123652380 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 39071 | gtex_brain_putamen_basal |
| rs881375 | 9 | 123652898 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 38553 | gtex_brain_putamen_basal |
| rs10117059 | 9 | 123653477 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 37974 | gtex_brain_putamen_basal |
| rs6478486 | 9 | 123655329 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 36122 | gtex_brain_putamen_basal |
| rs7037140 | 9 | 123656327 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 35124 | gtex_brain_putamen_basal |
| rs1468671 | 9 | 123657502 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 33949 | gtex_brain_putamen_basal |
| rs7036935 | 9 | 123658088 | TRAF1 | ENSG00000056558.6 | 1.958E-6 | 0 | 33363 | gtex_brain_putamen_basal |
| rs1548783 | 9 | 123658639 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 32812 | gtex_brain_putamen_basal |
| rs1860823 | 9 | 123659304 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 32147 | gtex_brain_putamen_basal |
| rs1860824 | 9 | 123659339 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 32112 | gtex_brain_putamen_basal |
| rs7046108 | 9 | 123660339 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 31112 | gtex_brain_putamen_basal |
| rs7875829 | 9 | 123660398 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 31053 | gtex_brain_putamen_basal |
| rs10818484 | 9 | 123661194 | TRAF1 | ENSG00000056558.6 | 2.274E-7 | 0 | 30257 | gtex_brain_putamen_basal |
| rs7019401 | 9 | 123661306 | TRAF1 | ENSG00000056558.6 | 1.731E-8 | 0 | 30145 | gtex_brain_putamen_basal |
| rs7859805 | 9 | 123664123 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 27328 | gtex_brain_putamen_basal |
| rs2241003 | 9 | 123666777 | TRAF1 | ENSG00000056558.6 | 1.056E-6 | 0 | 24674 | gtex_brain_putamen_basal |
| rs10818485 | 9 | 123667816 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 23635 | gtex_brain_putamen_basal |
| rs10435843 | 9 | 123668033 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 23418 | gtex_brain_putamen_basal |
| rs10435844 | 9 | 123668199 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 23252 | gtex_brain_putamen_basal |
| rs4837799 | 9 | 123668326 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 23125 | gtex_brain_putamen_basal |
| rs7848332 | 9 | 123669496 | TRAF1 | ENSG00000056558.6 | 9.188E-7 | 0 | 21955 | gtex_brain_putamen_basal |
| rs7866003 | 9 | 123670505 | TRAF1 | ENSG00000056558.6 | 2.15E-7 | 0 | 20946 | gtex_brain_putamen_basal |
| rs10985087 | 9 | 123671089 | TRAF1 | ENSG00000056558.6 | 1.383E-7 | 0 | 20362 | gtex_brain_putamen_basal |
| rs2239657 | 9 | 123671520 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 19931 | gtex_brain_putamen_basal |
| rs2239658 | 9 | 123671837 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 19614 | gtex_brain_putamen_basal |
| rs6478487 | 9 | 123672777 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 18674 | gtex_brain_putamen_basal |
| rs2416805 | 9 | 123676482 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 14969 | gtex_brain_putamen_basal |
| rs758959 | 9 | 123676699 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 14752 | gtex_brain_putamen_basal |
| rs876445 | 9 | 123677102 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 14349 | gtex_brain_putamen_basal |
| rs2109895 | 9 | 123677827 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 13624 | gtex_brain_putamen_basal |
| rs2159778 | 9 | 123678129 | TRAF1 | ENSG00000056558.6 | 9.643E-7 | 0 | 13322 | gtex_brain_putamen_basal |
| rs1008381 | 9 | 123681116 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 10335 | gtex_brain_putamen_basal |
| rs1008382 | 9 | 123681255 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 10196 | gtex_brain_putamen_basal |
| rs2109896 | 9 | 123683106 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 8345 | gtex_brain_putamen_basal |
| rs1008383 | 9 | 123683579 | TRAF1 | ENSG00000056558.6 | 7.451E-8 | 0 | 7872 | gtex_brain_putamen_basal |
| rs7021206 | 9 | 123684157 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 7294 | gtex_brain_putamen_basal |
| rs1014529 | 9 | 123684943 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 6508 | gtex_brain_putamen_basal |
| rs1548784 | 9 | 123685280 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 6171 | gtex_brain_putamen_basal |
| rs1930780 | 9 | 123686219 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 5232 | gtex_brain_putamen_basal |
| rs7033753 | 9 | 123686862 | TRAF1 | ENSG00000056558.6 | 4.466E-7 | 0 | 4589 | gtex_brain_putamen_basal |
| rs7034492 | 9 | 123687211 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 4240 | gtex_brain_putamen_basal |
| rs7034653 | 9 | 123687372 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 4079 | gtex_brain_putamen_basal |
| rs1930781 | 9 | 123687834 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 3617 | gtex_brain_putamen_basal |
| rs2416806 | 9 | 123690292 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | 1159 | gtex_brain_putamen_basal |
| rs3761849 | 9 | 123690957 | TRAF1 | ENSG00000056558.6 | 5.789E-7 | 0 | 494 | gtex_brain_putamen_basal |
| rs7028641 | 9 | 123691141 | TRAF1 | ENSG00000056558.6 | 7.393E-8 | 0 | 310 | gtex_brain_putamen_basal |
| rs1930786 | 9 | 123691469 | TRAF1 | ENSG00000056558.6 | 3.722E-8 | 0 | -18 | gtex_brain_putamen_basal |
| rs7864019 | 9 | 123692868 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | -1417 | gtex_brain_putamen_basal |
| rs10739579 | 9 | 123694788 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | -3337 | gtex_brain_putamen_basal |
| rs7031096 | 9 | 123694942 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | -3491 | gtex_brain_putamen_basal |
| rs10739580 | 9 | 123695282 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | -3831 | gtex_brain_putamen_basal |
| rs397768469 | 9 | 123695470 | TRAF1 | ENSG00000056558.6 | 2.912E-7 | 0 | -4019 | gtex_brain_putamen_basal |
| rs10733648 | 9 | 123700779 | TRAF1 | ENSG00000056558.6 | 1.381E-7 | 0 | -9328 | gtex_brain_putamen_basal |
| rs2416807 | 9 | 123704138 | TRAF1 | ENSG00000056558.6 | 8.395E-8 | 0 | -12687 | gtex_brain_putamen_basal |
| rs201413579 | 9 | 123704554 | TRAF1 | ENSG00000056558.6 | 8.383E-8 | 0 | -13103 | gtex_brain_putamen_basal |
| rs4837804 | 9 | 123705304 | TRAF1 | ENSG00000056558.6 | 1.388E-7 | 0 | -13853 | gtex_brain_putamen_basal |
| rs7039505 | 9 | 123705945 | TRAF1 | ENSG00000056558.6 | 1.582E-7 | 0 | -14494 | gtex_brain_putamen_basal |
| rs6478492 | 9 | 123706147 | TRAF1 | ENSG00000056558.6 | 2.716E-7 | 0 | -14696 | gtex_brain_putamen_basal |
| rs2900180 | 9 | 123706382 | TRAF1 | ENSG00000056558.6 | 1.227E-7 | 0 | -14931 | gtex_brain_putamen_basal |
| rs398087550 | 9 | 123708286 | TRAF1 | ENSG00000056558.6 | 1.737E-7 | 0 | -16835 | gtex_brain_putamen_basal |
| rs2075049 | 9 | 123715496 | TRAF1 | ENSG00000056558.6 | 4.056E-6 | 0 | -24045 | gtex_brain_putamen_basal |
| rs10818491 | 9 | 123723351 | TRAF1 | ENSG00000056558.6 | 4.036E-6 | 0 | -31900 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
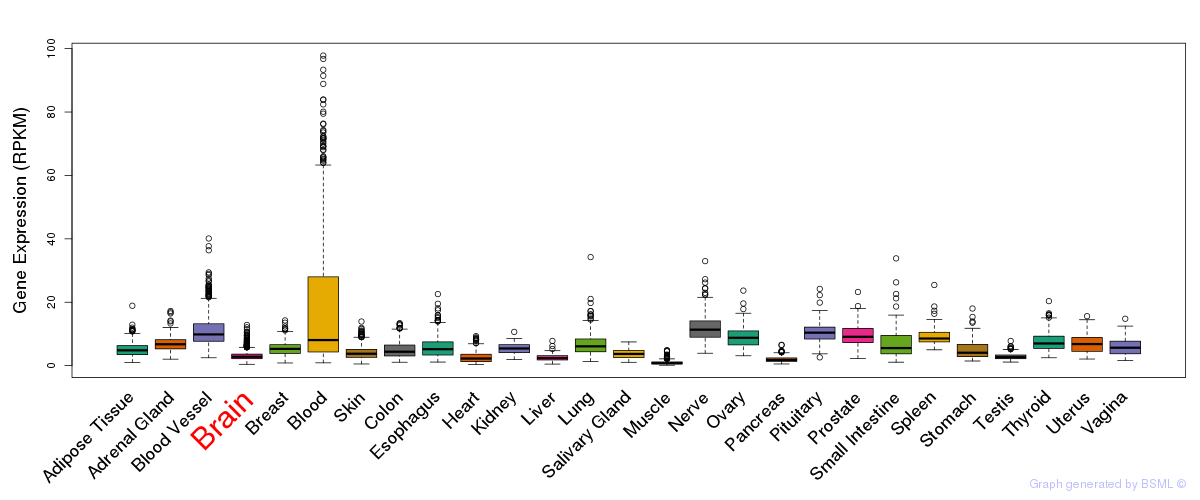
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 9162022 |10809768 | |
| GO:0008270 | zinc ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006461 | protein complex assembly | TAS | 8069916 | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0042981 | regulation of apoptosis | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | TAS | 8069916 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| APIP | APIP2 | CGI-29 | MMRP19 | dJ179L10.2 | APAF1 interacting protein | - | HPRD | 14743216 |
| BIRC2 | API1 | HIAP2 | Hiap-2 | MIHB | RNF48 | cIAP1 | baculoviral IAP repeat-containing 2 | c-IAP1 interacts with TRAF1. | BIND | 11907583 |
| BIRC2 | API1 | HIAP2 | Hiap-2 | MIHB | RNF48 | cIAP1 | baculoviral IAP repeat-containing 2 | - | HPRD,BioGRID | 9384571 |11907583 |
| BIRC3 | AIP1 | API2 | CIAP2 | HAIP1 | HIAP1 | MALT2 | MIHC | RNF49 | baculoviral IAP repeat-containing 3 | - | HPRD,BioGRID | 11907583 |
| BIRC3 | AIP1 | API2 | CIAP2 | HAIP1 | HIAP1 | MALT2 | MIHC | RNF49 | baculoviral IAP repeat-containing 3 | c-IAP2 interacts with TRAF1. | BIND | 11907583 |
| CASP10 | ALPS2 | FLICE2 | MCH4 | caspase 10, apoptosis-related cysteine peptidase | Biochemical Activity | BioGRID | 11098060 |
| CASP3 | CPP32 | CPP32B | SCA-1 | caspase 3, apoptosis-related cysteine peptidase | - | HPRD | 11098060 |
| CASP6 | MCH2 | caspase 6, apoptosis-related cysteine peptidase | Biochemical Activity | BioGRID | 11098060 |
| CASP8 | ALPS2B | CAP4 | FLICE | FLJ17672 | MACH | MCH5 | MGC78473 | caspase 8, apoptosis-related cysteine peptidase | Affinity Capture-Western Biochemical Activity | BioGRID | 11098060 |12887920 |
| CD27 | MGC20393 | S152 | T14 | TNFRSF7 | Tp55 | CD27 molecule | Affinity Capture-Western | BioGRID | 9794406 |
| CD40 | Bp50 | CDW40 | MGC9013 | TNFRSF5 | p50 | CD40 molecule, TNF receptor superfamily member 5 | - | HPRD | 7527023 |10411888 |
| CFLAR | CASH | CASP8AP1 | CLARP | Casper | FLAME | FLAME-1 | FLAME1 | FLIP | I-FLICE | MRIT | c-FLIP | c-FLIPL | c-FLIPR | c-FLIPS | CASP8 and FADD-like apoptosis regulator | - | HPRD,BioGRID | 9208847 |
| HIVEP3 | FLJ16752 | KBP-1 | KBP1 | KIAA1555 | KRC | SHN3 | Schnurri-3 | ZAS3 | ZNF40C | human immunodeficiency virus type I enhancer binding protein 3 | - | HPRD,BioGRID | 11804591 |
| HSPA8 | HSC54 | HSC70 | HSC71 | HSP71 | HSP73 | HSPA10 | LAP1 | MGC131511 | MGC29929 | NIP71 | heat shock 70kDa protein 8 | - | HPRD | 14743216 |
| LTBR | CD18 | D12S370 | LT-BETA-R | TNF-R-III | TNFCR | TNFR-RP | TNFR2-RP | TNFRSF3 | lymphotoxin beta receptor (TNFR superfamily, member 3) | Reconstituted Complex | BioGRID | 9162022 |
| MAP3K14 | FTDCR1B | HS | HSNIK | NIK | mitogen-activated protein kinase kinase kinase 14 | - | HPRD,BioGRID | 11278268 |
| MAP3K14 | FTDCR1B | HS | HSNIK | NIK | mitogen-activated protein kinase kinase kinase 14 | NIK interacts with TRAF1. This interaction was modeled on a demonstrated interaction between human NIK and TRAF1 from an unspecified species. | BIND | 9275204 |
| MAP3K5 | ASK1 | MAPKKK5 | MEKK5 | mitogen-activated protein kinase kinase kinase 5 | Affinity Capture-Western | BioGRID | 9774977 |
| MAP6 | FLJ41346 | KIAA1878 | MTAP6 | N-STOP | STOP | microtubule-associated protein 6 | in vitro | BioGRID | 11279055 |
| NGFR | CD271 | Gp80-LNGFR | TNFRSF16 | p75(NTR) | p75NTR | nerve growth factor receptor (TNFR superfamily, member 16) | Affinity Capture-Western | BioGRID | 10514511 |
| RIPK1 | FLJ39204 | RIP | RIP1 | receptor (TNFRSF)-interacting serine-threonine kinase 1 | - | HPRD,BioGRID | 8612133 |
| RIPK2 | CARD3 | CARDIAK | CCK | GIG30 | RICK | RIP2 | receptor-interacting serine-threonine kinase 2 | - | HPRD,BioGRID | 9705938 |
| RPS3 | FLJ26283 | FLJ27450 | MGC87870 | ribosomal protein S3 | - | HPRD | 14743216 |
| SPOP | TEF2 | speckle-type POZ protein | - | HPRD | 11279055 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | Affinity Capture-Western | BioGRID | 10635328 |
| TANK | I-TRAF | TRAF2 | TRAF family member-associated NFKB activator | - | HPRD,BioGRID | 8710854 |
| TNFAIP3 | A20 | MGC104522 | MGC138687 | MGC138688 | OTUD7C | TNFA1P2 | tumor necrosis factor, alpha-induced protein 3 | - | HPRD,BioGRID | 8692885 |
| TNFRSF11A | CD265 | FEO | LOH18CR1 | ODFR | OFE | OPTB7 | OSTS | PDB2 | RANK | TRANCER | tumor necrosis factor receptor superfamily, member 11a, NFKB activator | - | HPRD,BioGRID | 9774460 |9852070 |10025951 |
| TNFRSF12A | CD266 | FN14 | TWEAKR | tumor necrosis factor receptor superfamily, member 12A | - | HPRD,BioGRID | 11728344 |
| TNFRSF14 | ATAR | HVEA | HVEM | LIGHTR | TR2 | tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) | - | HPRD,BioGRID | 9162022 |
| TNFRSF17 | BCM | BCMA | CD269 | tumor necrosis factor receptor superfamily, member 17 | - | HPRD,BioGRID | 10903733 |
| TNFRSF18 | AITR | GITR | GITR-D | tumor necrosis factor receptor superfamily, member 18 | - | HPRD,BioGRID | 10037686 |
| TNFRSF19 | TAJ | TAJ-alpha | TRADE | TROY | tumor necrosis factor receptor superfamily, member 19 | - | HPRD,BioGRID | 10809768 |
| TNFRSF1A | CD120a | FPF | MGC19588 | TBP1 | TNF-R | TNF-R-I | TNF-R55 | TNFAR | TNFR1 | TNFR55 | TNFR60 | p55 | p55-R | p60 | tumor necrosis factor receptor superfamily, member 1A | - | HPRD,BioGRID | 8565075 |
| TNFRSF1B | CD120b | TBPII | TNF-R-II | TNF-R75 | TNFBR | TNFR1B | TNFR2 | TNFR80 | p75 | p75TNFR | tumor necrosis factor receptor superfamily, member 1B | - | HPRD | 8069916 |
| TNFRSF1B | CD120b | TBPII | TNF-R-II | TNF-R75 | TNFBR | TNFR1B | TNFR2 | TNFR80 | p75 | p75TNFR | tumor necrosis factor receptor superfamily, member 1B | Reconstituted Complex | BioGRID | 9162022 |
| TNFRSF8 | CD30 | D1S166E | KI-1 | tumor necrosis factor receptor superfamily, member 8 | - | HPRD,BioGRID | 8627180 |8943059 |9168896|8943059 |
| TNFRSF8 | CD30 | D1S166E | KI-1 | tumor necrosis factor receptor superfamily, member 8 | - | HPRD | 8943059 |
| TNFRSF8 | CD30 | D1S166E | KI-1 | tumor necrosis factor receptor superfamily, member 8 | TRAF1 interacts with CD30. | BIND | 8662842 |
| TNFRSF9 | 4-1BB | CD137 | CDw137 | ILA | MGC2172 | tumor necrosis factor receptor superfamily, member 9 | - | HPRD,BioGRID | 9464265 |
| TNFSF9 | 4-1BB-L | CD137L | tumor necrosis factor (ligand) superfamily, member 9 | - | HPRD,BioGRID | 9418902 |9607925 |
| TRADD | Hs.89862 | MGC11078 | TNFRSF1A-associated via death domain | - | HPRD,BioGRID | 8565075 |
| TRAF2 | MGC:45012 | TRAP | TRAP3 | TNF receptor-associated factor 2 | TRAF2 interacts with TRAF1. This interaction was modeled on a demonstrated interaction between human TRAF2 and mouse TRAF1. | BIND | 8702708 |
| TRAF2 | MGC:45012 | TRAP | TRAP3 | TNF receptor-associated factor 2 | - | HPRD,BioGRID | 8702708 |
| TRAIP | RNF206 | TRIP | TRAF interacting protein | - | HPRD,BioGRID | 9104814 |
| TRIM37 | KIAA0898 | MUL | POB1 | TEF3 | tripartite motif-containing 37 | - | HPRD,BioGRID | 11279055 |
| TUBA3C | TUBA2 | bA408E5.3 | tubulin, alpha 3c | - | HPRD | 14743216 |
| USP7 | HAUSP | TEF1 | ubiquitin specific peptidase 7 (herpes virus-associated) | - | HPRD,BioGRID | 11279055 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG SMALL CELL LUNG CANCER | 84 | 67 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HIVNEF PATHWAY | 58 | 43 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TNFR2 PATHWAY | 18 | 15 | All SZGR 2.0 genes in this pathway |
| PID CD40 PATHWAY | 31 | 26 | All SZGR 2.0 genes in this pathway |
| PID TNF PATHWAY | 46 | 33 | All SZGR 2.0 genes in this pathway |
| PID HIV NEF PATHWAY | 35 | 26 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP | 579 | 346 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE UP | 485 | 293 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA DN | 104 | 59 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 6HR DN | 41 | 29 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA UP | 544 | 308 | All SZGR 2.0 genes in this pathway |
| DUTTA APOPTOSIS VIA NFKB | 33 | 25 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B | 392 | 251 | All SZGR 2.0 genes in this pathway |
| SCHEIDEREIT IKK INTERACTING PROTEINS | 58 | 45 | All SZGR 2.0 genes in this pathway |
| DIRMEIER LMP1 RESPONSE EARLY | 66 | 48 | All SZGR 2.0 genes in this pathway |
| DIRMEIER LMP1 RESPONSE LATE UP | 57 | 41 | All SZGR 2.0 genes in this pathway |
| TSAI RESPONSE TO IONIZING RADIATION | 149 | 101 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| SCIAN INVERSED TARGETS OF TP53 AND TP73 DN | 31 | 23 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS DN | 411 | 249 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO IKK INHIBITOR AND TNF UP | 223 | 140 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| GOLDRATH IMMUNE MEMORY | 65 | 42 | All SZGR 2.0 genes in this pathway |
| GALINDO IMMUNE RESPONSE TO ENTEROTOXIN | 85 | 67 | All SZGR 2.0 genes in this pathway |
| BASSO CD40 SIGNALING UP | 101 | 76 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL LONG TERM | 302 | 191 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 24HR DN | 33 | 21 | All SZGR 2.0 genes in this pathway |
| GEISS RESPONSE TO DSRNA UP | 38 | 29 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 48HR DN | 161 | 105 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN | 409 | 268 | All SZGR 2.0 genes in this pathway |
| RAY TUMORIGENESIS BY ERBB2 CDC25A DN | 159 | 105 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA UP | 259 | 185 | All SZGR 2.0 genes in this pathway |
| VILIMAS NOTCH1 TARGETS UP | 52 | 41 | All SZGR 2.0 genes in this pathway |
| LINDSTEDT DENDRITIC CELL MATURATION B | 53 | 36 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP | 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 DN | 245 | 150 | All SZGR 2.0 genes in this pathway |
| VANASSE BCL2 TARGETS DN | 74 | 50 | All SZGR 2.0 genes in this pathway |
| HINATA NFKB TARGETS KERATINOCYTE UP | 91 | 63 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER UP | 288 | 168 | All SZGR 2.0 genes in this pathway |
| TIAN TNF SIGNALING VIA NFKB | 28 | 21 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP | 307 | 182 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS UP | 217 | 131 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS GROUP2 | 60 | 38 | All SZGR 2.0 genes in this pathway |
| WANG TNF TARGETS | 24 | 17 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38 | 337 | 236 | All SZGR 2.0 genes in this pathway |