Gene Page: TTN
Summary ?
| GeneID | 7273 |
| Symbol | TTN |
| Synonyms | CMD1G|CMH9|CMPD4|EOMFC|HMERF|LGMD2J|MYLK5|TMD |
| Description | titin |
| Reference | MIM:188840|HGNC:HGNC:12403|Ensembl:ENSG00000155657|HPRD:01787|Vega:OTTHUMG00000154448 |
| Gene type | protein-coding |
| Map location | 2q31 |
| Pascal p-value | 0.175 |
| Fetal beta | -1.551 |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| DNM:McCarthy_2014 | Whole Exome Sequencing analysis | Whole exome sequencing of 57 trios with sporadic or familial schizophrenia. | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0863 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| TTN | chr2 | 179417060 | G | A | NM_003319 | p.L21124L | synonymous SNV | Schizophrenia | DNM:McCarthy_2014 | ||
| TTN | chr2 | 179638834 | C | T | NM_001256850 NM_001256850 NM_001267550 NM_001267550 NM_003319 NM_003319 NM_133378 NM_133378 NM_133379 NM_133379 NM_133432 NM_133432 NM_133437 NM_133437 | . p.2354R>H . p.2354R>H . p.2308R>H . p.2354R>H . p.2354R>H . p.2308R>H . p.2308R>H | splice missense splice missense splice missense splice missense splice missense splice missense splice missense | Schizophrenia | DNM:Fromer_2014 |
Section II. Transcriptome annotation
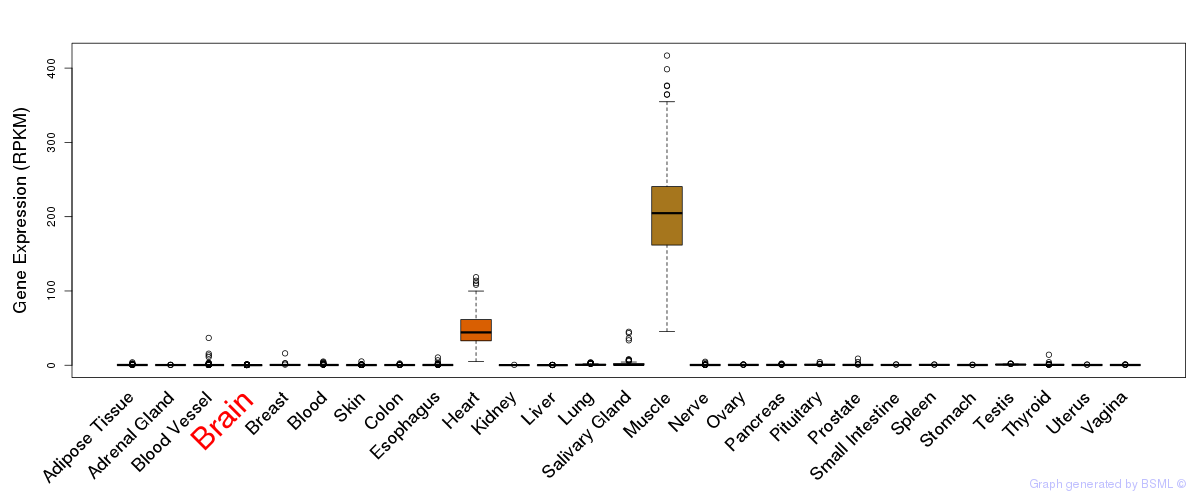
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TRAP1 | 0.88 | 0.87 |
| CFL1 | 0.87 | 0.88 |
| PPP2R1A | 0.87 | 0.87 |
| DCTN2 | 0.86 | 0.88 |
| ARPC1A | 0.86 | 0.86 |
| KAT5 | 0.86 | 0.85 |
| PSMD3 | 0.85 | 0.84 |
| CCT5 | 0.84 | 0.86 |
| AKR1B1 | 0.84 | 0.85 |
| NOC2L | 0.84 | 0.84 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.27 | -0.76 | -0.76 |
| AF347015.31 | -0.75 | -0.74 |
| AF347015.8 | -0.74 | -0.73 |
| AF347015.33 | -0.73 | -0.74 |
| MT-CO2 | -0.73 | -0.71 |
| AF347015.21 | -0.73 | -0.76 |
| MT-CYB | -0.73 | -0.73 |
| AF347015.2 | -0.71 | -0.71 |
| AF347015.15 | -0.71 | -0.71 |
| MT-ATP8 | -0.68 | -0.75 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005089 | Rho guanyl-nucleotide exchange factor activity | IEA | - | |
| GO:0004601 | peroxidase activity | IEA | - | |
| GO:0005516 | calmodulin binding | TAS | 10481174 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0005524 | ATP binding | TAS | 9804419 | |
| GO:0004674 | protein serine/threonine kinase activity | IDA | 9804419 | |
| GO:0004674 | protein serine/threonine kinase activity | IEA | - | |
| GO:0004713 | protein tyrosine kinase activity | IEA | - | |
| GO:0004197 | cysteine-type endopeptidase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0020037 | heme binding | IEA | - | |
| GO:0008307 | structural constituent of muscle | TAS | 7569978 | |
| GO:0017022 | myosin binding | TAS | 11717165 | |
| GO:0042802 | identical protein binding | IPI | 9804419 | |
| GO:0051393 | alpha-actinin binding | TAS | 10481174 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0006508 | proteolysis | IEA | - | |
| GO:0005975 | carbohydrate metabolic process | IEA | - | |
| GO:0007067 | mitosis | TAS | 10481174 | |
| GO:0006979 | response to oxidative stress | IEA | - | |
| GO:0006941 | striated muscle contraction | TAS | 7569978 | |
| GO:0035023 | regulation of Rho protein signal transduction | IEA | - | |
| GO:0030239 | myofibril assembly | IMP | 9804419 | |
| GO:0046777 | protein amino acid autophosphorylation | IMP | 9804419 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000794 | condensed nuclear chromosome | TAS | 10481174 | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0030017 | sarcomere | TAS | 7569978 | |
| GO:0030018 | Z disc | IDA | 9501083 | |
| GO:0030018 | Z disc | TAS | 8937992 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTN1 | FLJ40884 | actinin, alpha 1 | - | HPRD,BioGRID | 11305911 |11846417 |
| ANK1 | ANK | SPH1 | SPH2 | ankyrin 1, erythrocytic | - | HPRD,BioGRID | 12444090 |
| ANKRD1 | ALRP | C-193 | CARP | CVARP | MCARP | bA320F15.2 | ankyrin repeat domain 1 (cardiac muscle) | - | HPRD,BioGRID | 14583192 |
| ANKRD2 | ARPP | MGC104314 | ankyrin repeat domain 2 (stretch responsive muscle) | - | HPRD,BioGRID | 14583192 |
| ANKRD23 | DARP | FLJ32449 | MARP3 | MGC129593 | ankyrin repeat domain 23 | - | HPRD,BioGRID | 14583192 |
| ASF1B | CIA-II | FLJ10604 | ASF1 anti-silencing function 1 homolog B (S. cerevisiae) | Affinity Capture-MS | BioGRID | 17353931 |
| CALM1 | CALML2 | CAMI | DD132 | PHKD | calmodulin 1 (phosphorylase kinase, delta) | - | HPRD | 11178895 |
| CAPN3 | CANP3 | CANPL3 | LGMD2 | LGMD2A | MGC10767 | MGC11121 | MGC14344 | MGC4403 | nCL-1 | p94 | calpain 3, (p94) | - | HPRD | 8537379 |9642272|9185618 |9763216 |10987085 |
| CAPN3 | CANP3 | CANPL3 | LGMD2 | LGMD2A | MGC10767 | MGC11121 | MGC14344 | MGC4403 | nCL-1 | p94 | calpain 3, (p94) | - | HPRD,BioGRID | 8537379 |9642272 |
| FHL2 | AAG11 | DRAL | SLIM3 | four and a half LIM domains 2 | An unspecified isoform of FHL2 (DRAL) interacts with TTN (titin). | BIND | 12432079 |
| FHL2 | AAG11 | DRAL | SLIM3 | four and a half LIM domains 2 | - | HPRD,BioGRID | 12432079 |
| KY | FLJ33207 | kyphoscoliosis peptidase | KY interacts with an unspecified isoform of TTN. This interaction was modeled on a demonstrated interaction between mouse KY and human TTN. | BIND | 15385448 |
| MYBPC3 | CMH4 | DKFZp779E1762 | FHC | MYBP-C | myosin binding protein C, cardiac | - | HPRD,BioGRID | 8631348 |12202917 |
| MYOM1 | MGC134946 | MGC134947 | SKELEMIN | myomesin 1, 185kDa | - | HPRD | 7588733 |
| MYOM2 | TTNAP | myomesin (M-protein) 2, 165kDa | - | HPRD | 7505783 |
| MYPN | MYOP | myopalladin | Two-hybrid | BioGRID | 14583192 |
| NEB | DKFZp686C1456 | FLJ11505 | FLJ36536 | FLJ39568 | FLJ39584 | NEB177D | NEM2 | nebulin | - | HPRD | 11851340 |12482578 |
| OBSCN | DKFZp666E245 | FLJ14124 | KIAA1556 | KIAA1639 | MGC120409 | MGC120410 | MGC120411 | MGC120412 | MGC138590 | UNC89 | obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF | - | HPRD,BioGRID | 11448995 |
| PUF60 | FIR | FLJ31379 | RoBPI | SIAHBP1 | poly-U binding splicing factor 60KDa | Affinity Capture-MS | BioGRID | 17353931 |
| SQSTM1 | A170 | OSIL | PDB3 | ZIP3 | p60 | p62 | p62B | sequestosome 1 | Titin interacts with p62. This interaction was modelled on a demonstrated interaction between human Titin and rat p62. | BIND | 15802564 |
| TCAP | CMD1N | LGMD2G | T-cap | TELE | telethonin | titin-cap (telethonin) | - | HPRD,BioGRID | 9645487 |9804419 |9817758 |12446666 |
| TCAP | CMD1N | LGMD2G | T-cap | TELE | telethonin | titin-cap (telethonin) | Titin interacts with telethonin. This interaction was modelled on a demonstrated interaction between human Titin and telethonin from unspecified species. | BIND | 15802564 |
| TRIM63 | FLJ32380 | IRF | MURF1 | MURF2 | RNF28 | SMRZ | tripartite motif-containing 63 | Reconstituted Complex Two-hybrid | BioGRID | 11243782 |
| TRIM63 | FLJ32380 | IRF | MURF1 | MURF2 | RNF28 | SMRZ | tripartite motif-containing 63 | - | HPRD | 11927605 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG HYPERTROPHIC CARDIOMYOPATHY HCM | 85 | 65 | All SZGR 2.0 genes in this pathway |
| KEGG DILATED CARDIOMYOPATHY | 92 | 68 | All SZGR 2.0 genes in this pathway |
| DAVICIONI TARGETS OF PAX FOXO1 FUSIONS DN | 68 | 49 | All SZGR 2.0 genes in this pathway |
| HOOI ST7 TARGETS DN | 123 | 78 | All SZGR 2.0 genes in this pathway |
| AKL HTLV1 INFECTION DN | 68 | 41 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA PROGRESSION RISK | 74 | 44 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B | 392 | 251 | All SZGR 2.0 genes in this pathway |
| KORKOLA YOLK SAC TUMOR | 62 | 33 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK HIGH VS LOW DN | 32 | 21 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS DN | 124 | 79 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER F | 54 | 32 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO DN | 200 | 112 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB TARGETS | 74 | 41 | All SZGR 2.0 genes in this pathway |
| SHEPARD CRUSH AND BURN MUTANT DN | 185 | 111 | All SZGR 2.0 genes in this pathway |
| CHIARETTI ACUTE LYMPHOBLASTIC LEUKEMIA ZAP70 | 67 | 33 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| KUNINGER IGF1 VS PDGFB TARGETS UP | 82 | 51 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC UP | 202 | 115 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS UP | 170 | 107 | All SZGR 2.0 genes in this pathway |
| DAIRKEE CANCER PRONE RESPONSE BPA E2 | 118 | 65 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K27ME3 UP | 295 | 149 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE DN | 264 | 159 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP | 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF DN | 235 | 144 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 DN | 245 | 150 | All SZGR 2.0 genes in this pathway |
| GUTIERREZ CHRONIC LYMPHOCYTIC LEUKEMIA DN | 56 | 39 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE EARLY LATE | 317 | 190 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-101 | 205 | 211 | 1A | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-144 | 204 | 211 | 1A,m8 | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-17-5p/20/93.mr/106/519.d | 530 | 536 | 1A | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-208 | 538 | 544 | m8 | hsa-miR-208 | AUAAGACGAGCAAAAAGCUUGU |
| miR-24 | 830 | 836 | 1A | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
| miR-433-3p | 574 | 580 | m8 | hsa-miR-433brain | AUCAUGAUGGGCUCCUCGGUGU |
| miR-451 | 224 | 231 | 1A,m8 | hsa-miR-451brain | AAACCGUUACCAUUACUGAGUUU |
| miR-493-5p | 198 | 204 | m8 | hsa-miR-493-5p | UUGUACAUGGUAGGCUUUCAUU |
| miR-542-3p | 560 | 566 | 1A | hsa-miR-542-3p | UGUGACAGAUUGAUAACUGAAA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.