Gene Page: VDAC2
Summary ?
| GeneID | 7417 |
| Symbol | VDAC2 |
| Synonyms | POR |
| Description | voltage dependent anion channel 2 |
| Reference | MIM:193245|HGNC:HGNC:12672|Ensembl:ENSG00000165637|HPRD:01904|Vega:OTTHUMG00000018517 |
| Gene type | protein-coding |
| Map location | 10q22 |
| Pascal p-value | 0.652 |
| Sherlock p-value | 0.119 |
| Fetal beta | 0.168 |
| DMG | 1 (# studies) |
| eGene | Frontal Cortex BA9 |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_clathrin G2Cdb.human_mitochondria G2Cdb.human_Synaptosome G2Cdb.human_TAP-PSD-95-CORE G2Cdb.humanNRC G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg11906176 | 10 | 76970489 | VDAC2 | 1.338E-4 | -0.369 | 0.031 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
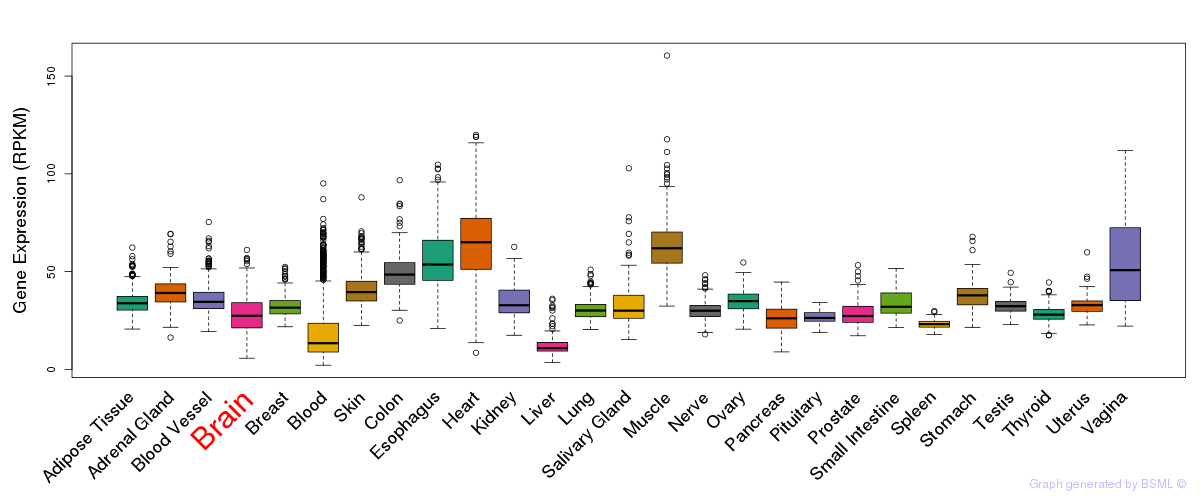
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CALCIUM SIGNALING PATHWAY | 178 | 134 | All SZGR 2.0 genes in this pathway |
| KEGG PARKINSONS DISEASE | 133 | 78 | All SZGR 2.0 genes in this pathway |
| KEGG HUNTINGTONS DISEASE | 185 | 109 | All SZGR 2.0 genes in this pathway |
| DITTMER PTHLH TARGETS UP | 112 | 68 | All SZGR 2.0 genes in this pathway |
| GALLUZZI PERMEABILIZE MITOCHONDRIA | 43 | 31 | All SZGR 2.0 genes in this pathway |
| GALLUZZI PREVENT MITOCHONDIAL PERMEABILIZATION | 22 | 16 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION DN | 517 | 309 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL AND BRAIN QTL TRANS | 185 | 114 | All SZGR 2.0 genes in this pathway |
| GOLDRATH HOMEOSTATIC PROLIFERATION | 171 | 102 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| KAYO CALORIE RESTRICTION MUSCLE DN | 87 | 59 | All SZGR 2.0 genes in this pathway |
| MAHAJAN RESPONSE TO IL1A DN | 76 | 57 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE DN | 123 | 76 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION NEOCORTEX DN | 88 | 58 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C1 | 72 | 45 | All SZGR 2.0 genes in this pathway |
| LEIN PONS MARKERS | 89 | 59 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS UP | 388 | 234 | All SZGR 2.0 genes in this pathway |
| WONG MITOCHONDRIA GENE MODULE | 217 | 122 | All SZGR 2.0 genes in this pathway |
| BRUECKNER TARGETS OF MIRLET7A3 UP | 111 | 69 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| NUTT GBM VS AO GLIOMA DN | 45 | 22 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| OKAMOTO LIVER CANCER MULTICENTRIC OCCURRENCE UP | 25 | 15 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| GUTIERREZ MULTIPLE MYELOMA UP | 35 | 24 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 10 | 69 | 38 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |