Gene Page: DNALI1
Summary ?
| GeneID | 7802 |
| Symbol | DNALI1 |
| Synonyms | P28|dJ423B22.5|hp28 |
| Description | dynein axonemal light intermediate chain 1 |
| Reference | MIM:602135|HGNC:HGNC:14353|Ensembl:ENSG00000163879|HPRD:03681|Vega:OTTHUMG00000004222 |
| Gene type | protein-coding |
| Map location | 1p35.1 |
| Pascal p-value | 0.39 |
| Sherlock p-value | 1.792E-4 |
| Fetal beta | 1.265 |
| eGene | Cerebellar Hemisphere Cerebellum Cortex Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17109968 | chr1 | 54633542 | DNALI1 | 7802 | 0 | trans | ||
| rs12476453 | chr2 | 4140214 | DNALI1 | 7802 | 0.1 | trans | ||
| rs4953981 | chr2 | 133216078 | DNALI1 | 7802 | 0.17 | trans | ||
| rs9369420 | chr6 | 9486466 | DNALI1 | 7802 | 0.14 | trans | ||
| rs16910520 | chr9 | 124088239 | DNALI1 | 7802 | 0.16 | trans | ||
| rs2289069 | chr9 | 124088730 | DNALI1 | 7802 | 0.16 | trans | ||
| rs2164298 | 0 | DNALI1 | 7802 | 0.16 | trans | |||
| rs3747847 | chr9 | 124111627 | DNALI1 | 7802 | 0.16 | trans | ||
| rs6478513 | chr9 | 124238226 | DNALI1 | 7802 | 0.12 | trans | ||
| rs2037311 | chr11 | 45873447 | DNALI1 | 7802 | 0.19 | trans | ||
| rs9300053 | chr11 | 45878875 | DNALI1 | 7802 | 0.06 | trans | ||
| rs12281674 | chr11 | 45899961 | DNALI1 | 7802 | 0.06 | trans | ||
| rs10746188 | chr12 | 81718315 | DNALI1 | 7802 | 0.12 | trans | ||
| snp_a-2238348 | 0 | DNALI1 | 7802 | 0.12 | trans | |||
| rs16962554 | chr19 | 29590544 | DNALI1 | 7802 | 0.13 | trans |
Section II. Transcriptome annotation
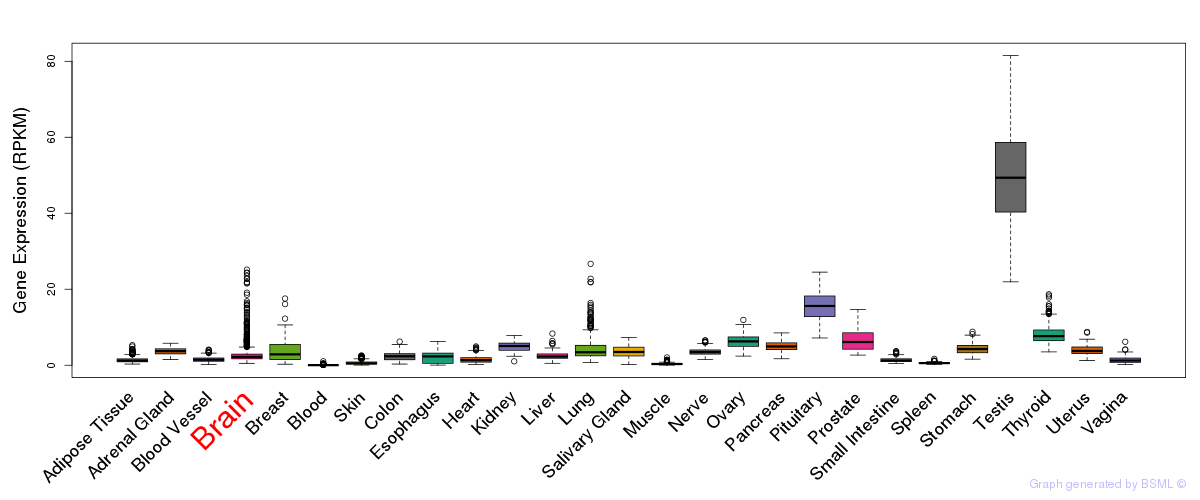
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG HUNTINGTONS DISEASE | 185 | 109 | All SZGR 2.0 genes in this pathway |
| KIM RESPONSE TO TSA AND DECITABINE UP | 129 | 73 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA DN | 349 | 157 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL UP | 380 | 215 | All SZGR 2.0 genes in this pathway |
| DOANE BREAST CANCER ESR1 UP | 112 | 72 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS LUMINAL | 326 | 213 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| LIEN BREAST CARCINOMA METAPLASTIC VS DUCTAL DN | 114 | 58 | All SZGR 2.0 genes in this pathway |
| YANG BREAST CANCER ESR1 BULK UP | 27 | 15 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 2 | 473 | 224 | All SZGR 2.0 genes in this pathway |
| LEIN CHOROID PLEXUS MARKERS | 103 | 61 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY GAMMA AND UV RADIATION | 88 | 65 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE UP | 226 | 164 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BRAIN DN | 85 | 58 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE UP | 97 | 61 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN LUNG DN | 37 | 22 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN LIVER DN | 10 | 9 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B UP | 172 | 109 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER ESR1 UP | 167 | 99 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER BRCA1 DN | 44 | 28 | All SZGR 2.0 genes in this pathway |
| KESHELAVA MULTIPLE DRUG RESISTANCE | 88 | 56 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS DN | 306 | 191 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC ICP WITH H3K4ME3 | 445 | 257 | All SZGR 2.0 genes in this pathway |