Gene Page: MIS12
Summary ?
| GeneID | 79003 |
| Symbol | MIS12 |
| Synonyms | 2510025F08Rik|KNTC2AP|MTW1|hMis12 |
| Description | MIS12 kinetochore complex component |
| Reference | MIM:609178|HGNC:HGNC:24967|Ensembl:ENSG00000167842|HPRD:12379|Vega:OTTHUMG00000102042 |
| Gene type | protein-coding |
| Map location | 17p13.2 |
| Pascal p-value | 0.183 |
| Sherlock p-value | 0.613 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg19775406 | 17 | 5390252 | MIS12 | 2.83E-10 | -0.018 | 6.59E-7 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7213696 | chr17 | 78721170 | MIS12 | 79003 | 0.2 | trans | ||
| rs5963536 | chrX | 38613824 | MIS12 | 79003 | 0.16 | trans |
Section II. Transcriptome annotation
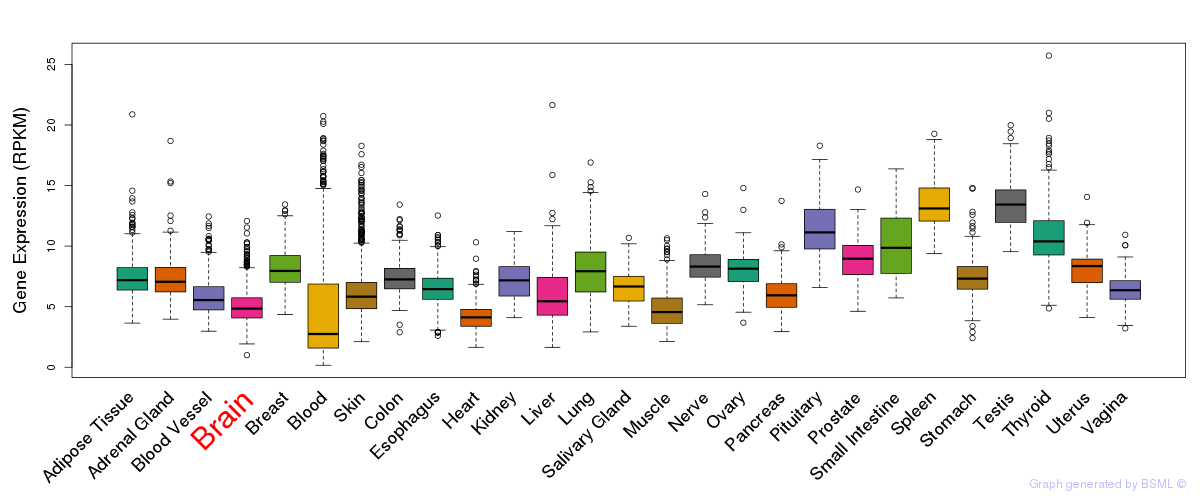
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CASC5 | AF15Q14 | D40 | KIAA1570 | KNL1 | cancer susceptibility candidate 5 | - | HPRD,BioGRID | 15502821 |
| CBX3 | HECH | HP1-GAMMA | HP1Hs-gamma | chromobox homolog 3 (HP1 gamma homolog, Drosophila) | - | HPRD,BioGRID | 15502821 |
| CBX3 | HECH | HP1-GAMMA | HP1Hs-gamma | chromobox homolog 3 (HP1 gamma homolog, Drosophila) | Mis12 interacts with HP1-gamma. | BIND | 15502821 |
| CBX5 | HP1 | HP1A | chromobox homolog 5 (HP1 alpha homolog, Drosophila) | - | HPRD,BioGRID | 15502821 |
| CBX5 | HP1 | HP1A | chromobox homolog 5 (HP1 alpha homolog, Drosophila) | Mis12 interacts with HP1-alpha. | BIND | 15502821 |
| DSN1 | C20orf172 | FLJ13346 | MGC32987 | MIS13 | dJ469A13.2 | DSN1, MIND kinetochore complex component, homolog (S. cerevisiae) | - | HPRD,BioGRID | 15502821 |
| DSN1 | C20orf172 | FLJ13346 | MGC32987 | MIS13 | dJ469A13.2 | DSN1, MIND kinetochore complex component, homolog (S. cerevisiae) | Mis12 interacts with c20orf172. | BIND | 15502821 |
| NDC80 | HEC | HEC1 | KNTC2 | TID3 | hsNDC80 | NDC80 homolog, kinetochore complex component (S. cerevisiae) | - | HPRD,BioGRID | 15502821 |
| NSL1 | C1orf48 | DC8 | DKFZp566O1646 | MIS14 | NSL1, MIND kinetochore complex component, homolog (S. cerevisiae) | Mis12 interacts with DC8. | BIND | 15502821 |
| NSL1 | C1orf48 | DC8 | DKFZp566O1646 | MIS14 | NSL1, MIND kinetochore complex component, homolog (S. cerevisiae) | - | HPRD,BioGRID | 15502821 |
| NUF2 | CDCA1 | NUF2R | NUF2, NDC80 kinetochore complex component, homolog (S. cerevisiae) | Affinity Capture-MS | BioGRID | 15371340 |
| PMF1 | - | polyamine-modulated factor 1 | - | HPRD,BioGRID | 15502821 |
| SPC24 | FLJ90806 | SPBC24 | SPC24, NDC80 kinetochore complex component, homolog (S. cerevisiae) | Affinity Capture-MS | BioGRID | 15371340 |
| SPC25 | AD024 | MGC22228 | SPBC25 | SPC25, NDC80 kinetochore complex component, homolog (S. cerevisiae) | Affinity Capture-MS | BioGRID | 15371340 |
| ZWINT | HZwint-1 | KNTC2AP | MGC117174 | ZWINT1 | ZW10 interactor | - | HPRD,BioGRID | 15502821 |
| ZWINT | HZwint-1 | KNTC2AP | MGC117174 | ZWINT1 | ZW10 interactor | Mis12 interacts with Zwint-1. | BIND | 15502821 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC M M G1 PHASES | 172 | 98 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPLICATION | 192 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC PROMETAPHASE | 87 | 51 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 5 6WK UP | 116 | 68 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER WITH LOH IN CHR9Q | 116 | 71 | All SZGR 2.0 genes in this pathway |
| GEORGES CELL CYCLE MIR192 TARGETS | 62 | 46 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| GALE APL WITH FLT3 MUTATED UP | 56 | 35 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |