Gene Page: PGBD5
Summary ?
| GeneID | 79605 |
| Symbol | PGBD5 |
| Synonyms | - |
| Description | piggyBac transposable element derived 5 |
| Reference | MIM:616791|HGNC:HGNC:19405|Ensembl:ENSG00000177614|HPRD:10148|Vega:OTTHUMG00000037759 |
| Gene type | protein-coding |
| Map location | 1q42.13 |
| Sherlock p-value | 0.141 |
| Fetal beta | -0.938 |
| DMG | 1 (# studies) |
| eGene | Cerebellar Hemisphere Cerebellum Meta |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 3 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg03347632 | 1 | 230457979 | PGBD5 | 1.182E-4 | 0.608 | 0.029 | DMG:Wockner_2014 |
| cg04095826 | 1 | 230457906 | PGBD5 | 1.771E-4 | 0.508 | 0.034 | DMG:Wockner_2014 |
| cg22875527 | 1 | 230492944 | PGBD5 | 5.525E-4 | 0.587 | 0.049 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
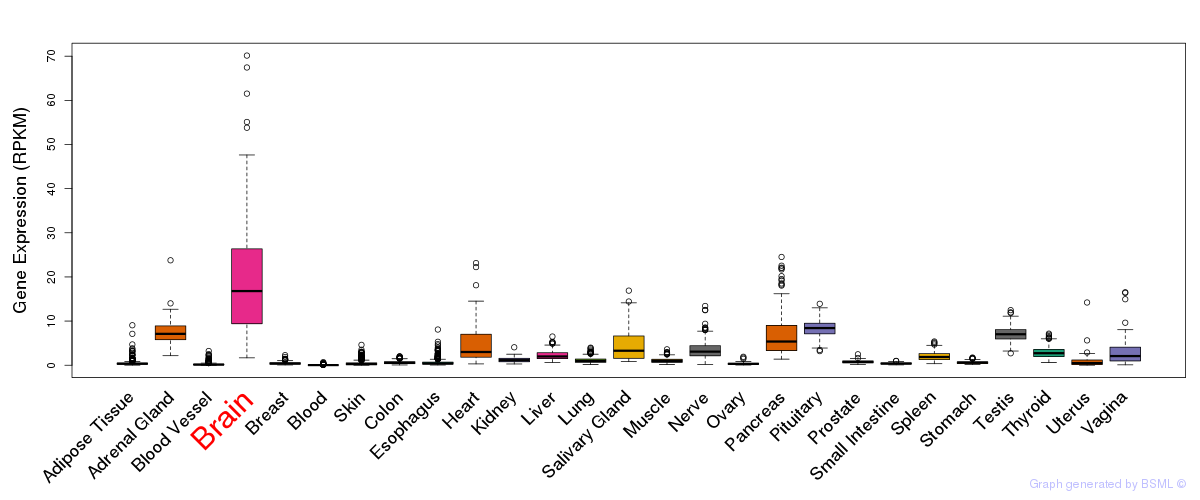
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MRPL22 | 0.76 | 0.71 |
| RPA3 | 0.75 | 0.66 |
| MRPS14 | 0.74 | 0.73 |
| NDUFA6 | 0.74 | 0.70 |
| TRMT112 | 0.74 | 0.67 |
| TOMM5 | 0.73 | 0.68 |
| ACN9 | 0.73 | 0.70 |
| MRPL13 | 0.73 | 0.67 |
| NSL1 | 0.73 | 0.73 |
| AK2 | 0.72 | 0.60 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.15 | -0.35 | -0.34 |
| AF347015.26 | -0.34 | -0.31 |
| AF347015.2 | -0.33 | -0.29 |
| AF347015.18 | -0.32 | -0.27 |
| AF347015.8 | -0.32 | -0.29 |
| MT-CYB | -0.32 | -0.31 |
| C9orf131 | -0.30 | -0.32 |
| BAI1 | -0.29 | -0.25 |
| AC100783.1 | -0.29 | -0.31 |
| MT-ATP8 | -0.29 | -0.25 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DAVICIONI MOLECULAR ARMS VS ERMS UP | 332 | 228 | All SZGR 2.0 genes in this pathway |
| DOANE BREAST CANCER ESR1 DN | 48 | 28 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS A DN | 90 | 61 | All SZGR 2.0 genes in this pathway |
| EBAUER TARGETS OF PAX3 FOXO1 FUSION DN | 48 | 31 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| AMIT SERUM RESPONSE 240 MCF10A | 57 | 36 | All SZGR 2.0 genes in this pathway |
| ROSS AML WITH PML RARA FUSION | 77 | 62 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE DN | 315 | 197 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 AND H3K27ME3 | 349 | 234 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K27ME3 | 341 | 243 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA NEURAL | 129 | 85 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS NOT VIA AKT1 UP | 211 | 131 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |