Gene Page: NARS2
Summary ?
| GeneID | 79731 |
| Symbol | NARS2 |
| Synonyms | DFNB94|SLM5|asnRS |
| Description | asparaginyl-tRNA synthetase 2, mitochondrial (putative) |
| Reference | MIM:612803|HGNC:HGNC:26274|Ensembl:ENSG00000137513|HPRD:08026|Vega:OTTHUMG00000166702 |
| Gene type | protein-coding |
| Map location | 11q14.1 |
| Pascal p-value | 0.216 |
| Sherlock p-value | 0.46 |
| Fetal beta | 0.857 |
| DMG | 1 (# studies) |
| eGene | Anterior cingulate cortex BA24 Cerebellar Hemisphere Cerebellum Cortex Frontal Cortex BA9 Hypothalamus Putamen basal ganglia Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg15143698 | 11 | 78286037 | NARS2 | 2.02E-8 | -0.015 | 6.99E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs9548306 | chr13 | 38876011 | NARS2 | 79731 | 0.01 | trans | ||
| rs7130503 | 11 | 78272226 | NARS2 | ENSG00000137513.5 | 1.221E-7 | 0 | 13693 | gtex_brain_ba24 |
| rs7101478 | 11 | 78272517 | NARS2 | ENSG00000137513.5 | 2.058E-9 | 0 | 13402 | gtex_brain_ba24 |
| rs9666103 | 11 | 78273948 | NARS2 | ENSG00000137513.5 | 4.645E-9 | 0 | 11971 | gtex_brain_ba24 |
| rs10899555 | 11 | 78274414 | NARS2 | ENSG00000137513.5 | 9.624E-7 | 0 | 11505 | gtex_brain_ba24 |
| rs10793333 | 11 | 78274652 | NARS2 | ENSG00000137513.5 | 2.048E-8 | 0 | 11267 | gtex_brain_ba24 |
| rs67659196 | 11 | 78274854 | NARS2 | ENSG00000137513.5 | 2.44E-9 | 0 | 11065 | gtex_brain_ba24 |
| rs12808344 | 11 | 78274964 | NARS2 | ENSG00000137513.5 | 5.464E-7 | 0 | 10955 | gtex_brain_ba24 |
| rs7129552 | 11 | 78275206 | NARS2 | ENSG00000137513.5 | 5.416E-9 | 0 | 10713 | gtex_brain_ba24 |
| rs6592794 | 11 | 78275743 | NARS2 | ENSG00000137513.5 | 5.491E-8 | 0 | 10176 | gtex_brain_ba24 |
| rs7936011 | 11 | 78275931 | NARS2 | ENSG00000137513.5 | 2.015E-7 | 0 | 9988 | gtex_brain_ba24 |
| rs7929656 | 11 | 78276416 | NARS2 | ENSG00000137513.5 | 2.18E-7 | 0 | 9503 | gtex_brain_ba24 |
| rs7940720 | 11 | 78276674 | NARS2 | ENSG00000137513.5 | 1.885E-8 | 0 | 9245 | gtex_brain_ba24 |
| rs10751296 | 11 | 78277277 | NARS2 | ENSG00000137513.5 | 2.497E-7 | 0 | 8642 | gtex_brain_ba24 |
| rs7108439 | 11 | 78277708 | NARS2 | ENSG00000137513.5 | 4.723E-7 | 0 | 8211 | gtex_brain_ba24 |
| rs201089218 | 11 | 78278013 | NARS2 | ENSG00000137513.5 | 3.721E-7 | 0 | 7906 | gtex_brain_ba24 |
| rs7937225 | 11 | 78278126 | NARS2 | ENSG00000137513.5 | 1.012E-8 | 0 | 7793 | gtex_brain_ba24 |
| rs10751297 | 11 | 78278567 | NARS2 | ENSG00000137513.5 | 4.215E-7 | 0 | 7352 | gtex_brain_ba24 |
| rs10793334 | 11 | 78278792 | NARS2 | ENSG00000137513.5 | 1.46E-8 | 0 | 7127 | gtex_brain_ba24 |
| rs10899556 | 11 | 78278986 | NARS2 | ENSG00000137513.5 | 5.168E-7 | 0 | 6933 | gtex_brain_ba24 |
| rs10793335 | 11 | 78279005 | NARS2 | ENSG00000137513.5 | 5.198E-7 | 0 | 6914 | gtex_brain_ba24 |
| rs10501429 | 11 | 78279790 | NARS2 | ENSG00000137513.5 | 1.884E-7 | 0 | 6129 | gtex_brain_ba24 |
| rs7942527 | 11 | 78280322 | NARS2 | ENSG00000137513.5 | 2.557E-8 | 0 | 5597 | gtex_brain_ba24 |
| rs7945812 | 11 | 78280509 | NARS2 | ENSG00000137513.5 | 2.42E-8 | 0 | 5410 | gtex_brain_ba24 |
| rs7927494 | 11 | 78280578 | NARS2 | ENSG00000137513.5 | 7.089E-7 | 0 | 5341 | gtex_brain_ba24 |
| rs7927503 | 11 | 78280606 | NARS2 | ENSG00000137513.5 | 2.018E-7 | 0 | 5313 | gtex_brain_ba24 |
| rs7124803 | 11 | 78280822 | NARS2 | ENSG00000137513.5 | 2.47E-8 | 0 | 5097 | gtex_brain_ba24 |
| rs10793336 | 11 | 78281867 | NARS2 | ENSG00000137513.5 | 6.652E-7 | 0 | 4052 | gtex_brain_ba24 |
| rs4945300 | 11 | 78282632 | NARS2 | ENSG00000137513.5 | 1.027E-7 | 0 | 3287 | gtex_brain_ba24 |
| rs4944210 | 11 | 78282878 | NARS2 | ENSG00000137513.5 | 6.2E-7 | 0 | 3041 | gtex_brain_ba24 |
| rs4945301 | 11 | 78283362 | NARS2 | ENSG00000137513.5 | 6.079E-7 | 0 | 2557 | gtex_brain_ba24 |
| rs4945302 | 11 | 78283454 | NARS2 | ENSG00000137513.5 | 7.079E-7 | 0 | 2465 | gtex_brain_ba24 |
| rs4945303 | 11 | 78283803 | NARS2 | ENSG00000137513.5 | 9.792E-8 | 0 | 2116 | gtex_brain_ba24 |
| rs7940507 | 11 | 78284453 | NARS2 | ENSG00000137513.5 | 2.266E-8 | 0 | 1466 | gtex_brain_ba24 |
| rs7937417 | 11 | 78284470 | NARS2 | ENSG00000137513.5 | 1.945E-8 | 0 | 1449 | gtex_brain_ba24 |
| rs4367973 | 11 | 78284545 | NARS2 | ENSG00000137513.5 | 9.603E-7 | 0 | 1374 | gtex_brain_ba24 |
| rs4457782 | 11 | 78284733 | NARS2 | ENSG00000137513.5 | 5.838E-7 | 0 | 1186 | gtex_brain_ba24 |
| rs4275650 | 11 | 78284822 | NARS2 | ENSG00000137513.5 | 5.819E-7 | 0 | 1097 | gtex_brain_ba24 |
| rs4277140 | 11 | 78285096 | NARS2 | ENSG00000137513.5 | 2.497E-7 | 0 | 823 | gtex_brain_ba24 |
| rs10793337 | 11 | 78285875 | NARS2 | ENSG00000137513.5 | 1.806E-9 | 0 | 44 | gtex_brain_ba24 |
| rs10736814 | 11 | 78286462 | NARS2 | ENSG00000137513.5 | 2.361E-7 | 0 | -543 | gtex_brain_ba24 |
| rs7952030 | 11 | 78270304 | NARS2 | ENSG00000137513.5 | 1.677E-6 | 0.01 | 15615 | gtex_brain_putamen_basal |
| rs7101478 | 11 | 78272517 | NARS2 | ENSG00000137513.5 | 9.601E-8 | 0.01 | 13402 | gtex_brain_putamen_basal |
| rs9666103 | 11 | 78273948 | NARS2 | ENSG00000137513.5 | 1.111E-6 | 0.01 | 11971 | gtex_brain_putamen_basal |
| rs10899555 | 11 | 78274414 | NARS2 | ENSG00000137513.5 | 8.26E-8 | 0.01 | 11505 | gtex_brain_putamen_basal |
| rs67659196 | 11 | 78274854 | NARS2 | ENSG00000137513.5 | 5.028E-7 | 0.01 | 11065 | gtex_brain_putamen_basal |
| rs7129552 | 11 | 78275206 | NARS2 | ENSG00000137513.5 | 7.302E-7 | 0.01 | 10713 | gtex_brain_putamen_basal |
| rs7936011 | 11 | 78275931 | NARS2 | ENSG00000137513.5 | 5.511E-7 | 0.01 | 9988 | gtex_brain_putamen_basal |
| rs7929656 | 11 | 78276416 | NARS2 | ENSG00000137513.5 | 3.988E-7 | 0.01 | 9503 | gtex_brain_putamen_basal |
| rs10751296 | 11 | 78277277 | NARS2 | ENSG00000137513.5 | 2.486E-7 | 0.01 | 8642 | gtex_brain_putamen_basal |
| rs7108439 | 11 | 78277708 | NARS2 | ENSG00000137513.5 | 1.669E-7 | 0.01 | 8211 | gtex_brain_putamen_basal |
| rs201089218 | 11 | 78278013 | NARS2 | ENSG00000137513.5 | 3.699E-7 | 0.01 | 7906 | gtex_brain_putamen_basal |
| rs7937225 | 11 | 78278126 | NARS2 | ENSG00000137513.5 | 1.262E-7 | 0.01 | 7793 | gtex_brain_putamen_basal |
| rs10751297 | 11 | 78278567 | NARS2 | ENSG00000137513.5 | 5.052E-7 | 0.01 | 7352 | gtex_brain_putamen_basal |
| rs10793334 | 11 | 78278792 | NARS2 | ENSG00000137513.5 | 1.869E-7 | 0.01 | 7127 | gtex_brain_putamen_basal |
| rs10899556 | 11 | 78278986 | NARS2 | ENSG00000137513.5 | 6.194E-7 | 0.01 | 6933 | gtex_brain_putamen_basal |
| rs10793335 | 11 | 78279005 | NARS2 | ENSG00000137513.5 | 6.237E-7 | 0.01 | 6914 | gtex_brain_putamen_basal |
| rs7942527 | 11 | 78280322 | NARS2 | ENSG00000137513.5 | 3.201E-7 | 0.01 | 5597 | gtex_brain_putamen_basal |
| rs7945812 | 11 | 78280509 | NARS2 | ENSG00000137513.5 | 3.039E-7 | 0.01 | 5410 | gtex_brain_putamen_basal |
| rs7927494 | 11 | 78280578 | NARS2 | ENSG00000137513.5 | 1.029E-6 | 0.01 | 5341 | gtex_brain_putamen_basal |
| rs7927503 | 11 | 78280606 | NARS2 | ENSG00000137513.5 | 3.69E-7 | 0.01 | 5313 | gtex_brain_putamen_basal |
| rs7124803 | 11 | 78280822 | NARS2 | ENSG00000137513.5 | 2.99E-7 | 0.01 | 5097 | gtex_brain_putamen_basal |
| rs10793336 | 11 | 78281867 | NARS2 | ENSG00000137513.5 | 8.424E-7 | 0.01 | 4052 | gtex_brain_putamen_basal |
| rs4945300 | 11 | 78282632 | NARS2 | ENSG00000137513.5 | 1.381E-6 | 0.01 | 3287 | gtex_brain_putamen_basal |
| rs4944210 | 11 | 78282878 | NARS2 | ENSG00000137513.5 | 7.778E-7 | 0.01 | 3041 | gtex_brain_putamen_basal |
| rs4945301 | 11 | 78283362 | NARS2 | ENSG00000137513.5 | 7.606E-7 | 0.01 | 2557 | gtex_brain_putamen_basal |
| rs4945302 | 11 | 78283454 | NARS2 | ENSG00000137513.5 | 1.172E-6 | 0.01 | 2465 | gtex_brain_putamen_basal |
| rs4945303 | 11 | 78283803 | NARS2 | ENSG00000137513.5 | 2.453E-7 | 0.01 | 2116 | gtex_brain_putamen_basal |
| rs7940507 | 11 | 78284453 | NARS2 | ENSG00000137513.5 | 2.492E-7 | 0.01 | 1466 | gtex_brain_putamen_basal |
| rs7937417 | 11 | 78284470 | NARS2 | ENSG00000137513.5 | 2.468E-7 | 0.01 | 1449 | gtex_brain_putamen_basal |
| rs4367973 | 11 | 78284545 | NARS2 | ENSG00000137513.5 | 7.834E-7 | 0.01 | 1374 | gtex_brain_putamen_basal |
| rs4457782 | 11 | 78284733 | NARS2 | ENSG00000137513.5 | 7.263E-7 | 0.01 | 1186 | gtex_brain_putamen_basal |
| rs4275650 | 11 | 78284822 | NARS2 | ENSG00000137513.5 | 7.236E-7 | 0.01 | 1097 | gtex_brain_putamen_basal |
| rs4277140 | 11 | 78285096 | NARS2 | ENSG00000137513.5 | 2.486E-7 | 0.01 | 823 | gtex_brain_putamen_basal |
| rs10793337 | 11 | 78285875 | NARS2 | ENSG00000137513.5 | 1.868E-7 | 0.01 | 44 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
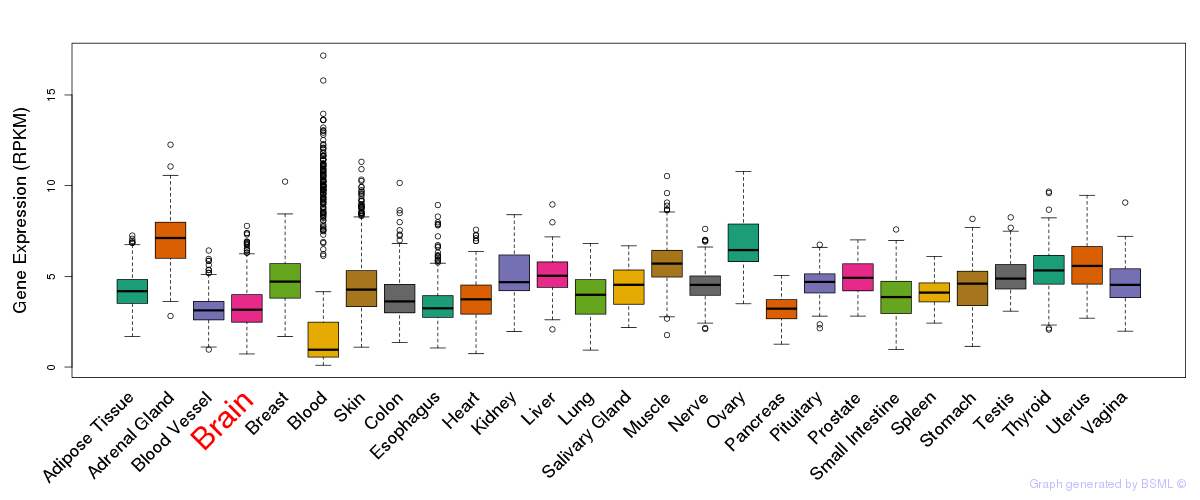
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG AMINOACYL TRNA BIOSYNTHESIS | 41 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | 21 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME TRNA AMINOACYLATION | 42 | 34 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| GROSS ELK3 TARGETS UP | 27 | 16 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 11Q12 Q14 AMPLICON | 158 | 93 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA DN | 289 | 166 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| WALLACE PROSTATE CANCER RACE DN | 88 | 42 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER UP | 288 | 168 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SURVIVAL DN | 113 | 76 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |