Gene Page: BAP1
Summary ?
| GeneID | 8314 |
| Symbol | BAP1 |
| Synonyms | HUCEP-13|UCHL2|hucep-6 |
| Description | BRCA1 associated protein 1 |
| Reference | MIM:603089|HGNC:HGNC:950|Ensembl:ENSG00000163930|HPRD:04366|Vega:OTTHUMG00000158392 |
| Gene type | protein-coding |
| Map location | 3p21.1 |
| Pascal p-value | 0.02 |
| Fetal beta | -0.604 |
| eGene | Cortex Myers' cis & trans Meta |
| Support | CompositeSet Darnell FMRP targets Chromatin Remodeling Genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| LK:YES | Genome-wide Association Study | This data set included 99 genes mapped to the 22 regions. The 24 leading SNPs were also included in CV:Ripke_2013 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1249912 | chr3 | 112390577 | BAP1 | 8314 | 0.15 | trans |
Section II. Transcriptome annotation
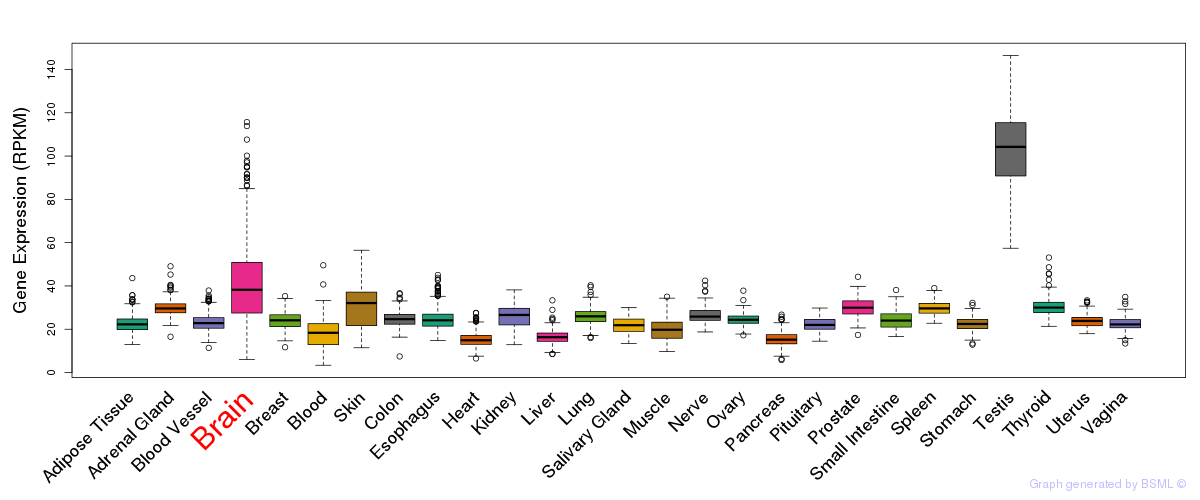
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MAP1LC3A | 0.85 | 0.88 |
| TM7SF2 | 0.80 | 0.78 |
| DEF8 | 0.80 | 0.85 |
| ABHD14A | 0.79 | 0.87 |
| WDR45 | 0.79 | 0.84 |
| SCO2 | 0.78 | 0.81 |
| CDK5 | 0.78 | 0.83 |
| SLC39A3 | 0.77 | 0.84 |
| MRPS12 | 0.77 | 0.85 |
| RPUSD3 | 0.77 | 0.86 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| EIF5B | -0.46 | -0.59 |
| NSBP1 | -0.44 | -0.54 |
| AF347015.18 | -0.43 | -0.37 |
| AC010300.1 | -0.40 | -0.53 |
| AF347015.26 | -0.39 | -0.30 |
| AC016705.1 | -0.39 | -0.33 |
| AF347015.2 | -0.38 | -0.26 |
| MT-ATP8 | -0.38 | -0.28 |
| Z83840.4 | -0.37 | -0.35 |
| AF347015.8 | -0.37 | -0.26 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| MEINHOLD OVARIAN CANCER LOW GRADE DN | 20 | 16 | All SZGR 2.0 genes in this pathway |
| PUJANA BREAST CANCER LIT INT NETWORK | 101 | 73 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS DN | 261 | 155 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER MUTATED FREQUENTLY | 12 | 10 | All SZGR 2.0 genes in this pathway |
| ZUCCHI METASTASIS DN | 44 | 29 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR UP | 240 | 152 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 16HR UP | 225 | 139 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE UP | 393 | 244 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY 4NQO OR UV | 63 | 44 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE UP | 226 | 164 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED | 351 | 238 | All SZGR 2.0 genes in this pathway |
| FONTAINE FOLLICULAR THYROID ADENOMA DN | 68 | 45 | All SZGR 2.0 genes in this pathway |
| FONTAINE THYROID TUMOR UNCERTAIN MALIGNANCY DN | 26 | 14 | All SZGR 2.0 genes in this pathway |
| FONTAINE PAPILLARY THYROID CARCINOMA DN | 80 | 53 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING DN | 225 | 124 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| YU BAP1 TARGETS | 29 | 21 | All SZGR 2.0 genes in this pathway |