Gene Page: CARS
Summary ?
| GeneID | 833 |
| Symbol | CARS |
| Synonyms | CARS1|CYSRS|MGC:11246 |
| Description | cysteinyl-tRNA synthetase |
| Reference | MIM:123859|HGNC:HGNC:1493|Ensembl:ENSG00000110619|HPRD:00464|Vega:OTTHUMG00000010927 |
| Gene type | protein-coding |
| Map location | 11p15.5 |
| Pascal p-value | 0.646 |
| Sherlock p-value | 0.592 |
| Fetal beta | -0.684 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Expression | Meta-analysis of gene expression | P value: 2.231 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg03336167 | 11 | 2930995 | CARS | 0.003 | -2.163 | DMG:vanEijk_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6048101 | chr20 | 22429986 | CARS | 833 | 0.11 | trans | ||
| rs4632416 | chr20 | 22437090 | CARS | 833 | 0.05 | trans | ||
| rs4459793 | chr20 | 22439811 | CARS | 833 | 0.07 | trans | ||
| rs6048112 | chr20 | 22442814 | CARS | 833 | 0.1 | trans |
Section II. Transcriptome annotation
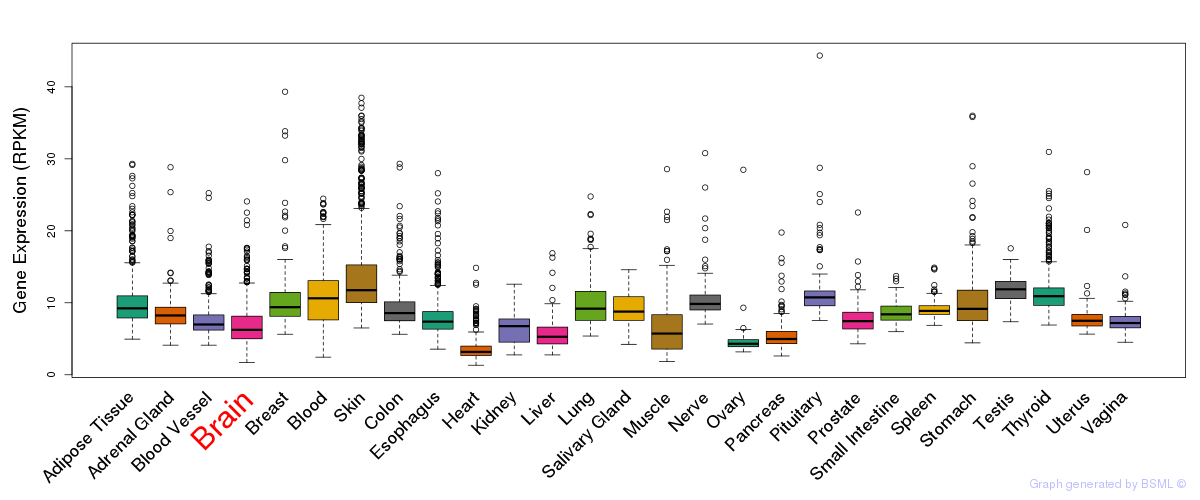
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| B2M | 0.71 | 0.74 |
| XXbac-BPG116M5.1 | 0.68 | 0.74 |
| VAMP5 | 0.68 | 0.72 |
| TM4SF1 | 0.68 | 0.70 |
| LST1 | 0.67 | 0.66 |
| HIGD1B | 0.66 | 0.68 |
| CFB | 0.66 | 0.73 |
| NMI | 0.65 | 0.69 |
| IGFBP7 | 0.65 | 0.68 |
| IFI27 | 0.64 | 0.64 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HNRNPUL2 | -0.58 | -0.57 |
| CDC2L2 | -0.57 | -0.57 |
| SUPT6H | -0.57 | -0.58 |
| PELP1 | -0.57 | -0.61 |
| HERC1 | -0.57 | -0.60 |
| NOC2L | -0.56 | -0.59 |
| SPTAN1 | -0.56 | -0.59 |
| SMARCC2 | -0.56 | -0.59 |
| MYO18A | -0.56 | -0.55 |
| NEURL4 | -0.56 | -0.59 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000049 | tRNA binding | IMP | 17303165 | |
| GO:0000049 | tRNA binding | NAS | 7987009 | |
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 10908348 | |
| GO:0005524 | ATP binding | IDA | 17303165 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004817 | cysteine-tRNA ligase activity | IDA | 10908348 |17303165 | |
| GO:0004817 | cysteine-tRNA ligase activity | IEA | - | |
| GO:0016874 | ligase activity | IEA | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0042803 | protein homodimerization activity | IMP | 17303165 | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006423 | cysteinyl-tRNA aminoacylation | IDA | 10908348 |17303165 | |
| GO:0006423 | cysteinyl-tRNA aminoacylation | IEA | - | |
| GO:0006412 | translation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG AMINOACYL TRNA BIOSYNTHESIS | 41 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOSOLIC TRNA AMINOACYLATION | 24 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME TRNA AMINOACYLATION | 42 | 34 | All SZGR 2.0 genes in this pathway |
| NAKAMURA CANCER MICROENVIRONMENT DN | 46 | 29 | All SZGR 2.0 genes in this pathway |
| LIU SOX4 TARGETS DN | 309 | 191 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP | 633 | 376 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| PROVENZANI METASTASIS UP | 194 | 112 | All SZGR 2.0 genes in this pathway |
| XU HGF SIGNALING NOT VIA AKT1 48HR UP | 35 | 20 | All SZGR 2.0 genes in this pathway |
| DIRMEIER LMP1 RESPONSE LATE UP | 57 | 41 | All SZGR 2.0 genes in this pathway |
| AMUNDSON RESPONSE TO ARSENITE | 217 | 143 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 DN | 156 | 106 | All SZGR 2.0 genes in this pathway |
| PENG LEUCINE DEPRIVATION UP | 142 | 93 | All SZGR 2.0 genes in this pathway |
| PENG GLUCOSE DEPRIVATION UP | 48 | 26 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA UP | 64 | 46 | All SZGR 2.0 genes in this pathway |
| ABE VEGFA TARGETS 2HR | 34 | 21 | All SZGR 2.0 genes in this pathway |
| LEE AGING CEREBELLUM DN | 86 | 66 | All SZGR 2.0 genes in this pathway |
| GAJATE RESPONSE TO TRABECTEDIN UP | 67 | 45 | All SZGR 2.0 genes in this pathway |
| TSENG IRS1 TARGETS UP | 113 | 71 | All SZGR 2.0 genes in this pathway |
| TSENG ADIPOGENIC POTENTIAL UP | 30 | 19 | All SZGR 2.0 genes in this pathway |
| KRIGE AMINO ACID DEPRIVATION | 29 | 20 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| PODAR RESPONSE TO ADAPHOSTIN UP | 147 | 98 | All SZGR 2.0 genes in this pathway |
| CHEN HOXA5 TARGETS 9HR UP | 223 | 132 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB UP | 245 | 159 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| CHAUHAN RESPONSE TO METHOXYESTRADIOL UP | 51 | 32 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED | 351 | 238 | All SZGR 2.0 genes in this pathway |
| GREGORY SYNTHETIC LETHAL WITH IMATINIB | 145 | 83 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38 | 337 | 236 | All SZGR 2.0 genes in this pathway |