Gene Page: STX7
Summary ?
| GeneID | 8417 |
| Symbol | STX7 |
| Synonyms | - |
| Description | syntaxin 7 |
| Reference | MIM:603217|HGNC:HGNC:11442|HPRD:04449| |
| Gene type | protein-coding |
| Map location | 6q23.1 |
| Pascal p-value | 0.073 |
| Sherlock p-value | 0.459 |
| eGene | Myers' cis & trans |
| Support | INTRACELLULAR TRAFFICKING G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_clathrin G2Cdb.human_Synaptosome |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs712785 | chr3 | 7485713 | STX7 | 8417 | 0.18 | trans |
Section II. Transcriptome annotation
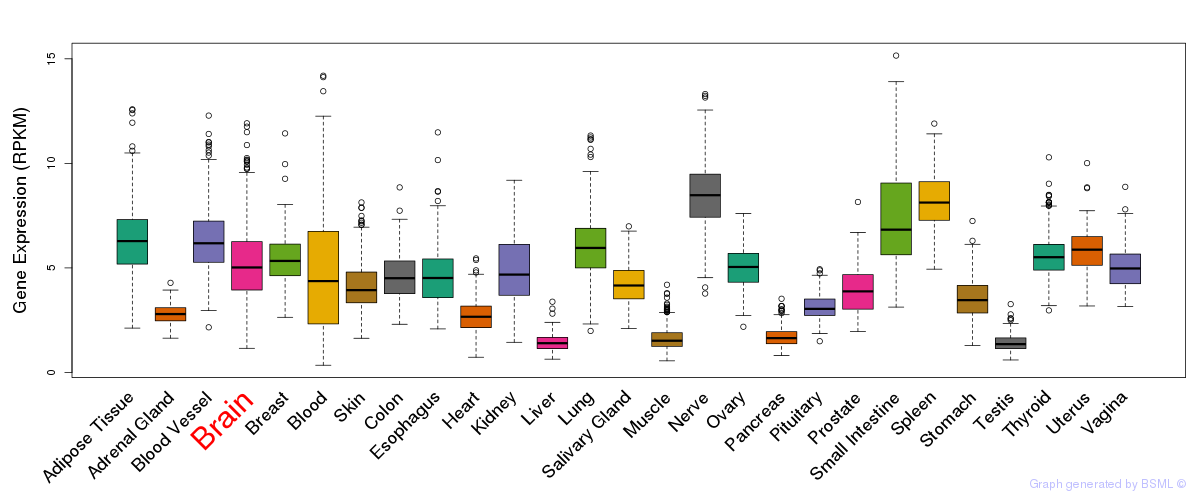
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HS3ST1 | 0.67 | 0.75 |
| KCNJ11 | 0.66 | 0.82 |
| SCRT1 | 0.66 | 0.79 |
| EPHB6 | 0.66 | 0.75 |
| OSBP2 | 0.65 | 0.81 |
| POU6F1 | 0.64 | 0.80 |
| KLC2 | 0.64 | 0.79 |
| IFFO1 | 0.64 | 0.79 |
| LRRC4 | 0.63 | 0.82 |
| ARHGEF11 | 0.63 | 0.84 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.46 | -0.67 |
| AF347015.31 | -0.46 | -0.61 |
| ACSF2 | -0.44 | -0.62 |
| MT-CO2 | -0.44 | -0.60 |
| S100A16 | -0.44 | -0.59 |
| AF347015.8 | -0.43 | -0.60 |
| AP002478.3 | -0.43 | -0.63 |
| AF347015.2 | -0.42 | -0.55 |
| AL139819.3 | -0.42 | -0.59 |
| DBI | -0.42 | -0.64 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005484 | SNAP receptor activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006886 | intracellular protein transport | IEA | - | |
| GO:0006892 | post-Golgi vesicle-mediated transport | TAS | 9358037 | |
| GO:0016192 | vesicle-mediated transport | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005768 | endosome | IEA | - | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0031901 | early endosome membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ENTPD2 | CD39L1 | NTPDase-2 | ectonucleoside triphosphate diphosphohydrolase 2 | - | HPRD | 11591653 |
| NAPA | SNAPA | N-ethylmaleimide-sensitive factor attachment protein, alpha | - | HPRD,BioGRID | 9417091 |
| SNAP29 | CEDNIK | FLJ21051 | SNAP-29 | synaptosomal-associated protein, 29kDa | - | HPRD,BioGRID | 9852078 |11423532 |
| STX6 | - | syntaxin 6 | - | HPRD,BioGRID | 11278762 |
| STX8 | CARB | syntaxin 8 | - | HPRD,BioGRID | 12175335 |
| VAMP7 | SYBL1 | TI-VAMP | VAMP-7 | vesicle-associated membrane protein 7 | in vitro in vivo | BioGRID | 11278762 |12175335 |
| VAMP7 | SYBL1 | TI-VAMP | VAMP-7 | vesicle-associated membrane protein 7 | - | HPRD | 11278762|12175335 |
| VAMP8 | EDB | vesicle-associated membrane protein 8 (endobrevin) | - | HPRD,BioGRID | 11278762 |
| VPS11 | END1 | PEP5 | RNF108 | hVPS11 | vacuolar protein sorting 11 homolog (S. cerevisiae) | VPS11 interacts with STX7 (Syntaxin-7). This interaction was modeled on a demonstrated interaction between human VPS11 and STX7 (Syntaxin-7) from an unspecified species. | BIND | 14623309 |
| VPS11 | END1 | PEP5 | RNF108 | hVPS11 | vacuolar protein sorting 11 homolog (S. cerevisiae) | - | HPRD,BioGRID | 11382755 |
| VPS16 | hVPS16 | vacuolar protein sorting 16 homolog (S. cerevisiae) | - | HPRD | 11382755 |
| VPS18 | KIAA1475 | PEP3 | vacuolar protein sorting 18 homolog (S. cerevisiae) | VPS18 interacts with STX7 (Syntaxin-7). This interaction was modeled on a demonstrated interaction between human VPS18 and STX7 (Syntaxin-7) from an unspecified species. | BIND | 14623309 |
| VPS18 | KIAA1475 | PEP3 | vacuolar protein sorting 18 homolog (S. cerevisiae) | - | HPRD,BioGRID | 11382755 |
| VTI1A | MVti1 | Vti1-rp2 | vesicle transport through interaction with t-SNAREs homolog 1A (yeast) | Affinity Capture-Western | BioGRID | 11101518 |
| VTI1B | VTI1 | VTI1-LIKE | VTI1L | VTI2 | vesicle transport through interaction with t-SNAREs homolog 1B (yeast) | - | HPRD,BioGRID | 12114520 |12175335 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG SNARE INTERACTIONS IN VESICULAR TRANSPORT | 38 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME BOTULINUM NEUROTOXICITY | 19 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | 17 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS UP | 332 | 228 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS DN | 431 | 263 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| WATTEL AUTONOMOUS THYROID ADENOMA UP | 73 | 47 | All SZGR 2.0 genes in this pathway |
| LIANG HEMATOPOIESIS STEM CELL NUMBER SMALL VS HUGE UP | 38 | 29 | All SZGR 2.0 genes in this pathway |
| PASQUALUCCI LYMPHOMA BY GC STAGE DN | 165 | 104 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS UP | 424 | 268 | All SZGR 2.0 genes in this pathway |
| KLEIN PRIMARY EFFUSION LYMPHOMA DN | 58 | 42 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D7 | 40 | 21 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE DN | 373 | 196 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE UP | 181 | 106 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP | 307 | 182 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS DURATION CORR DN | 146 | 90 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION UP | 271 | 165 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-361 | 163 | 169 | m8 | hsa-miR-361brain | UUAUCAGAAUCUCCAGGGGUAC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.