Gene Page: LINGO1
Summary ?
| GeneID | 84894 |
| Symbol | LINGO1 |
| Synonyms | LERN1|LRRN6A|UNQ201 |
| Description | leucine rich repeat and Ig domain containing 1 |
| Reference | MIM:609791|HGNC:HGNC:21205|Ensembl:ENSG00000169783|HPRD:14322|Vega:OTTHUMG00000172629 |
| Gene type | protein-coding |
| Map location | 15q24.3 |
| Pascal p-value | 0.067 |
| Sherlock p-value | 0.49 |
| Fetal beta | 0.702 |
| DMG | 1 (# studies) |
| eGene | Caudate basal ganglia Cerebellar Hemisphere Putamen basal ganglia |
| Support | INTRACELLULAR SIGNAL TRANSDUCTION CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg10107330 | 15 | 77922677 | LINGO1 | 1.26E-5 | 0.647 | 0.014 | DMG:Wockner_2014 |
| cg26884775 | 15 | 77906127 | LINGO1 | 4.059E-4 | 0.351 | 0.044 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2667781 | 15 | 77858619 | LINGO1 | ENSG00000169783.8 | 2.411E-6 | 0.04 | 254623 | gtex_brain_putamen_basal |
| rs2667782 | 15 | 77862296 | LINGO1 | ENSG00000169783.8 | 2.46E-6 | 0.04 | 250946 | gtex_brain_putamen_basal |
| rs2667783 | 15 | 77862529 | LINGO1 | ENSG00000169783.8 | 2.468E-6 | 0.04 | 250713 | gtex_brain_putamen_basal |
| rs2667775 | 15 | 77864802 | LINGO1 | ENSG00000169783.8 | 2.468E-6 | 0.04 | 248440 | gtex_brain_putamen_basal |
| rs2682921 | 15 | 77865459 | LINGO1 | ENSG00000169783.8 | 2.468E-6 | 0.04 | 247783 | gtex_brain_putamen_basal |
| rs939488 | 15 | 77867780 | LINGO1 | ENSG00000169783.8 | 2.721E-6 | 0.04 | 245462 | gtex_brain_putamen_basal |
| rs868297 | 15 | 77873769 | LINGO1 | ENSG00000169783.8 | 2.571E-6 | 0.04 | 239473 | gtex_brain_putamen_basal |
| rs2667769 | 15 | 77874877 | LINGO1 | ENSG00000169783.8 | 2.552E-6 | 0.04 | 238365 | gtex_brain_putamen_basal |
| rs2667768 | 15 | 77875117 | LINGO1 | ENSG00000169783.8 | 2.56E-6 | 0.04 | 238125 | gtex_brain_putamen_basal |
| rs2682911 | 15 | 77875591 | LINGO1 | ENSG00000169783.8 | 2.571E-6 | 0.04 | 237651 | gtex_brain_putamen_basal |
| rs2667767 | 15 | 77876031 | LINGO1 | ENSG00000169783.8 | 2.113E-6 | 0.04 | 237211 | gtex_brain_putamen_basal |
| rs867918 | 15 | 77882726 | LINGO1 | ENSG00000169783.8 | 2.593E-6 | 0.04 | 230516 | gtex_brain_putamen_basal |
| rs58600827 | 15 | 77884322 | LINGO1 | ENSG00000169783.8 | 2.583E-6 | 0.04 | 228920 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
General gene expression (GTEx)
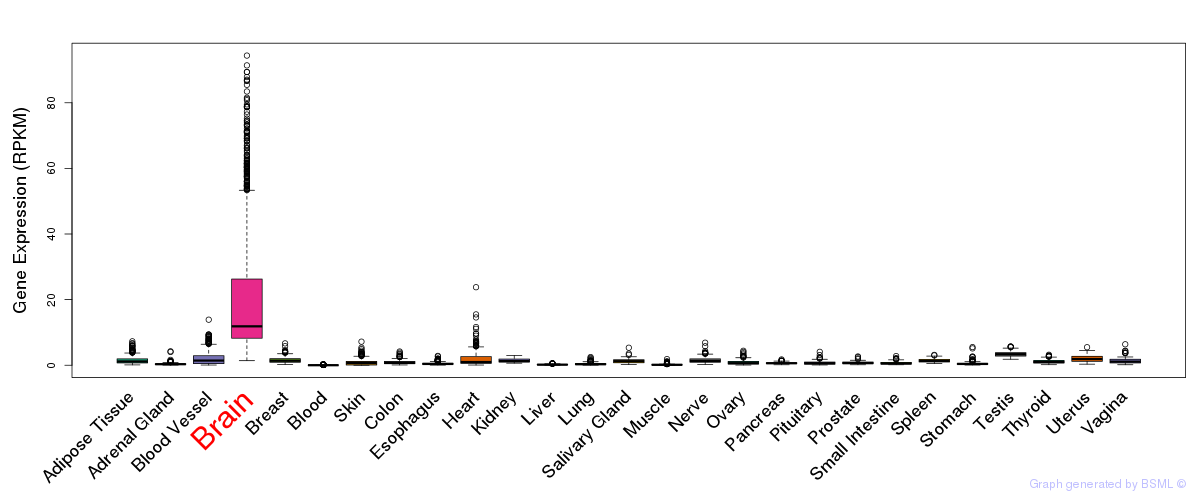
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID P75 NTR PATHWAY | 69 | 51 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | 81 | 61 | All SZGR 2.0 genes in this pathway |
| PEREZ TP63 TARGETS | 355 | 243 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH OCT4 TARGETS | 290 | 172 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH NOS TARGETS | 179 | 105 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION UP | 195 | 138 | All SZGR 2.0 genes in this pathway |
| STAEGE EWING FAMILY TUMOR | 33 | 22 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 2 | 473 | 224 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| YAMASHITA LIVER CANCER STEM CELL UP | 47 | 38 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |